1 Introduction
Yeasts are unicellular fungi that belong to different taxonomic groups. Among these, species of the Saccharomyces sensu stricto complex have been widely used by humans for preparing food and beverages. Despite the fact that bread and alcoholic beverage making are particularly ancient, the domestication of yeast has only recently been studied. Researchers have investigated the archaeological record, the process of fermentation, the history of wine yeast populations, species formation associated with beer or bread yeasts or the molecular signatures of human selection, etc. So far, no attempt has been made to put together all these pieces of information. In this review, we summarize the main results of this research and highlight the need for analyzing natural populations and for combining different approaches for a better understanding of yeast domestication.
2 Bread and alcohol beverages of the past: first traces of yeast domestication
It is only in 1860 that it was discovered that yeasts converted sugar into ethanol and carbon dioxide, a process called fermentation [1]. Although the peoples of ancient times had no understanding of this biochemical process, fermentation was used for thousands of years as an effective way to preserve the quality and safety of beverages and foods.
Archaeologists have found molecular evidence for the production of a fermented beverage dated back to 7000 BC from the Neolithic village of Jiahu in China [2]; similar evidence dated to 5400–5000 BC comes from the site of Hajji Firuz Tepe in the northern Zagros Mountains in Mesopotamia [3]. In Asia, fermented beverages were mainly made from rice. In ancient Egypt and Mesopotamia the most used beverages were those made from fruits (wine), from honey (mead) and from malted cereals (beer). Evidence of leavened bread was found in Ancient Egypt at the same time period [4].
According to what is believed today, grapes have been domesticated between the Black Sea and Iran during the 7000–4000 BC period [3,5–7]. The first evidence of wine making comes from the presence of tartaric acid in an ancient jar dated from 5400–5000 BC in the Neolithic site of Tepe in Mesopotamia [3] and from the remains for grape juice extraction from 5000 BC in the Neolithic site of Dikili Tash in Greece [8]. When wine was made from wild or cultivated grape remains to be elucidated but archaeological records suggest that wine was made as follows: grapes were hand picked and placed in a vat for traditional grape-treading, or in special wine presses. The resultant juice was captured in open jars, where the fermentation process took place. When ready, these jars were sealed and marked with the date, name of the vineyard and the person in charge of the wine. After aging, they had to be broken when it was time to decant the wine, and then poured into another jar. When the wine was ready to be served, it was poured into shallow vessels with a short stem. Wine was mainly drunk and offered to gods during religious ceremonies or was used in medicine [9] while beer was a popular drink.
Beer was first mentioned in the Mesopotamian “Epic of Gilgamesh”, one of the earliest works of literature. In the poem the “wild man”, Enkidu is given beer to drink. According to the poem, “he ate until he was full, drank seven pitchers of beer, his face glowed and he sang out with joy”. The ancient way of brewing can now be partly retrieved from tomb scenes, clay tablets as well as beer remains in pottery vessels [4,10]. Both emmer wheat (Triticum dicoccum Schübl.) and barley (Hordeum vulgare L.) were used for brewing, either separately or together. Beer was initially thought to have been made from yeasted dough: the dough was lightly baked and the resulting bread was crumbled and strained through a sieve with water [4,11]. However, the analysis of 1500–1300 BC old beer remains by electronic microscopy now suggests that instead beer was made from cooked and uncooked malt with water and the mixture was strained free of husk before inoculation with yeast [4]. Beer was either filtered or not, in which case it was drunk with a straw. Whether the yeast came from bread dough, the water of the Nile, or air inoculums remains to be elucidated [12].
Description of bread making and analysis of bread remains from ancient Egypt suggest an unsophisticated preparation. Bread was made of coarsely milled emmer wheat (Triticum dicoccum Schübl.) [4]. Analysis of Egyptian bread microstructure and starch from 1300–1500 BC remains suggest that the dough was moist rather than dry but kneading was not extensive [4]. Deliberately germinated grains, called malt, must have been used for some types of bread. Because emmer breads were dense it is difficult to observe yeast in ancient bread remains. However, yeasts were observed in some breads indicating that at least some leavened breads were made at that time [13].
It is expected that starting from those times, beverages and bread fermentation technologies expanded from Asia, Mesopotamia and Egypt throughout the Old World. While the origin and expansion of the process of leavened breads are poorly documented, the history of wine and beer making is better known (see for review [7]). The cultivation of grapevines and the production of wine spread all over the Mediterranean Sea towards Greece (5000 BC) [8], Italy (900 BC) [14], France (600 BC) [15], Northern Europe (100 AD) and much later to the Americas (1500 AD) [7,16]. Beer making diverged into two processes depending on the temperature and the yeast used during the fermentation steps. Ale beer technology, characterized by top-fermentation at 15–20 °C and followed in some cases by aging, was acquired from the Middle East by Germanic and Celtic tribes around the 1st century AD, whereas lager beer technology, characterized by bottom fermentation at 5–14 °C and a long period of maturation, appeared more recently during the Middle Ages in Europe [17,18].
In the Neolithic time, fermentations were probably initiated by naturally occurring yeasts, and exchanges of yeast between food and fermented beverages were likely. At what period of time human started to consciously add selected yeast in any of these processes is still unknown.
3 Commonly used yeast species for alcohol and bread fermentation
Among the fermenting microorganisms, yeasts are probably the most important group of eukaryotic microorganisms that have been used in production of fermented beverages and foods [19]. Yeasts are classically defined as single-celled fungi. Around 600 yeast species have currently been recorded and are widely distributed in diverse ecological niches and geographical areas. Here, we will focus on yeast species belonging to the Saccharomyces sensu stricto species complex which contains some of the most important species involved in alcoholic beverages as well as in bread fermentation (Fig. 1). These yeasts have a number of unique characters not found in other yeast genera [20]. While a majority of yeasts cannot grow in the absence of oxygen (aerobic yeasts), a majority of the Saccharomyces complex yeasts can survive without any oxygen by using the fermentation process [21–23]. As their name says, Saccharomyces sensu stricto yeasts species are specialized for growth on carbohydrates. In the absence of oxygen, they transform carbohydrates, usually six-carbon molecules such as glucose, into ethanol and CO2 via the fermentation process (Fig. 2). Interestingly, this can also occur in the presence of oxygen when there is a high concentration of carbohydrates because a “glucose repression” circuit represses the respiratory part in the presence of glucose [24]. The occurrence of fermentation under aerobic conditions is sometimes referred to as the Crabtree effect and the yeasts exhibiting this peculiarity are referred to as Crabtree-positive yeasts [22]. After depletion of glucose and accumulation of ethanol, the metabolism of Crabtree positive yeast changes: in presence of oxygen, respiration occurs and ethanol becomes a substrate that is degraded. The change of metabolism from the production of ethanol via fermentation to the consumption of ethanol via the respiration process is referred to as the diauxic shift. This shift requires several elements including the alcohol dehydrogenase enzymes (Adh). In S. cerevisiae, Adh1 has been shown to be predominantly responsible for the production of ethanol while Adh2 is expected to use preferentially ethanol as substrate. Reconstitution of the ADH ancestral gene [25] suggests that the ability to produce ethanol arose first and that the consumption of ethanol was then selected after the genome duplication estimated at 100 Mya [26,27]. Adh specialization would have occurred after the end of the cretaceous concomitant to the formation of fruits. Altogether these data revealed that Saccharomyces yeast species acquired their competitive advantage in rich environments such as alcoholic beverages and food by gaining the ability of producing high amounts of ethanol and tolerating it, as well as through their ability to grow in both anaerobic and aerobic conditions. Note that these innovations occurred far before Neolithic times or any domestication process.
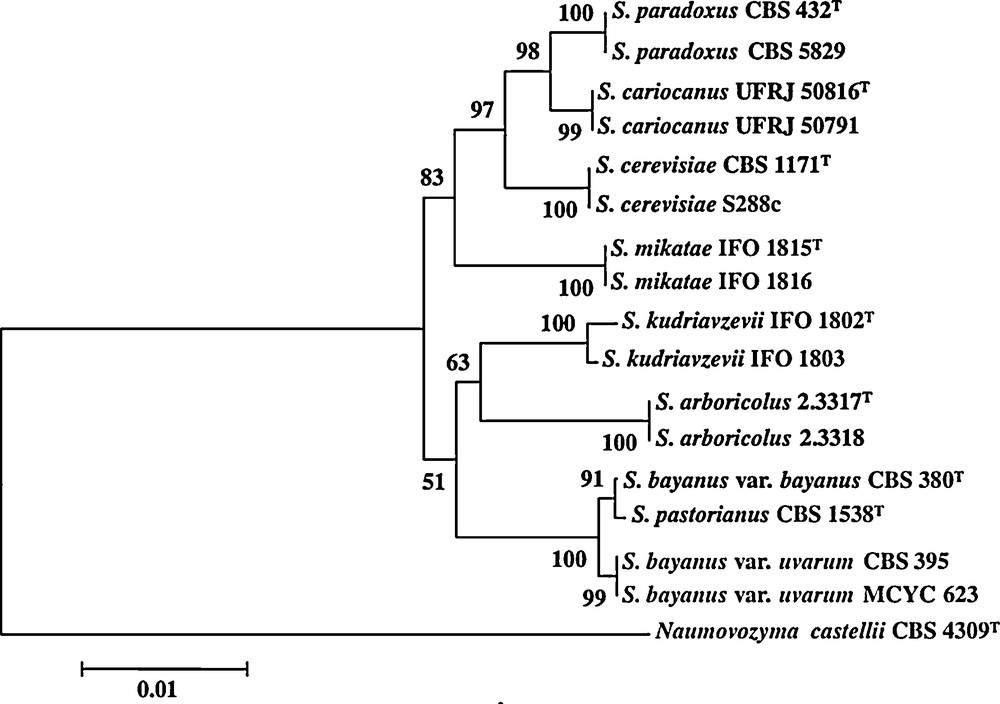
Clade of the Saccharomyces sensu stricto species complex.
From Naumov et al. 2010. The scale corresponds to a substitution rate of 10 bases per kb. Bootsrap values from 1000 replications are shown at each node when values are greater than 50%.
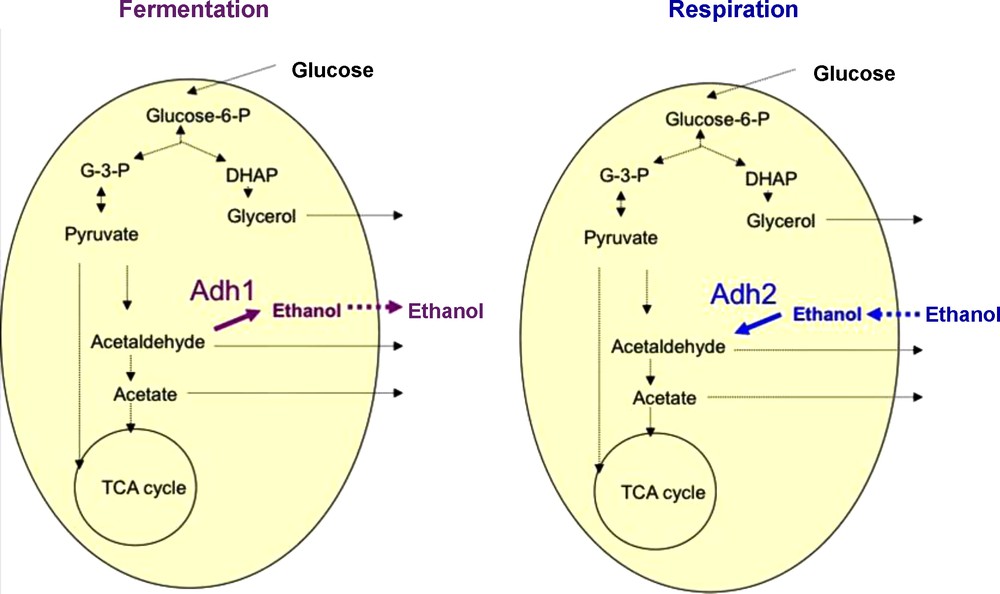
The make-accumulate-consume strategy of Saccharomyces yeasts. (Adh1: Alcohol dehydrogenase 1, Adh2: Alcohol dehydrogenase 2)
Within the Saccharomyces sensu stricto species complex, S. cariocanus, S. mikatae, S. paradoxus and S. kudriavzevii have been mostly found in natural environments and rarely in anthropic ones [28]. They are the other species of the complex that have been used extensively by humans for making beverages or bread. Nevertheless, the most commonly used species is S. cerevisiae that is considered to be the agent of wine, ale beer, sake, palm fermentation as well as the one of leavened bread [29–32]. The species S. bayanus is involved in lager beer fermentation [28,33] whereas S. uvarum has been isolated from wine and cider fermentations [28]. Hybridizations between species of the sensu stricto complex as well as variation of ploidy have occurred many times and have participated to the evolution of domesticated yeast [28,31].
4 S. cerevisiae genome diversity reflects human history and spread of grapevine production
Since the characterization of S. cerevisiae, numerous strains of this species have been isolated from beverages and food but only few have been found in nature, usually in soil or bark of oak trees. This has led to a controversy on whether S. cerevisiae was a domesticated species that arose from the action of humans or whether it appeared in nature and then was selected by human populations. Comparison of sets of S. cerevisiae domesticated and natural strains has revealed that the oldest lineages and most of the genetic variability occur in natural strains while domesticated strains harbored less diversity. This is in agreement with the hypothesis of an origin of the S. cerevisiae species in natural environments followed by human domestication [29].
Analysis of genetic diversity among a very large collection of S. cerevisiae coming from all over the world and from various ecological niches and fermentation processes (forest, fruits, clinical isolates, sake, wine, palm wine, beer, bread…) have recently revealed the evolutionary history of domesticated populations (Fig. 3). Yeast strains genetically clustered according to their ecological niches and type of fermentation rather than according to their geographical origins [29,32,34–38]. These results indicate that exchanges between niches or fermentation processes were not frequent enough to homogenize populations and suggested that independent domestication events occurred for sake, wine, palm wine, and beer making. The wide geographical distribution of some yeasts (beer, bread, cheese etc.) belonging to the same genetic group suggested that human participated widely to the rapid expansion of those yeast populations and fermentation processes. The pattern of diversity was also shaped by modern practices. For example, two groups of wine strains (522 Davies and Champagne strains groups, Fig. 3) contained closely genetically related industrial strains that had been extensively used all over the world. Bread strains behaved differently than beverage strains. They cluster on their own or belong to the wine cluster, suggesting they may have diverse origins.
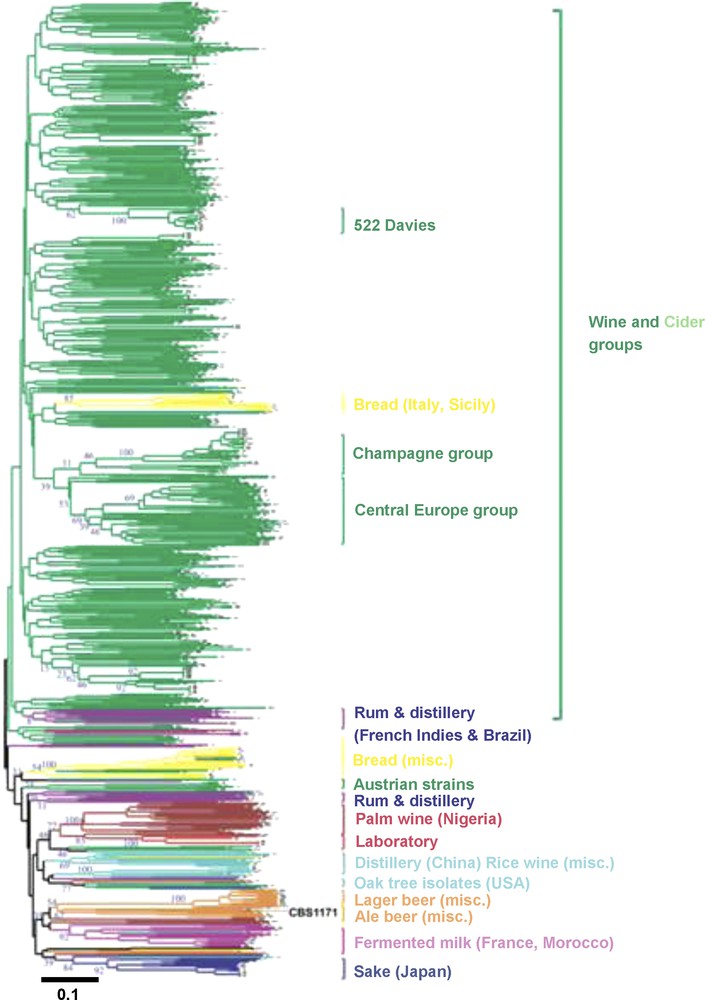
Neighbor-joining tree showing the clustering of 651 yeast strains isolated from different sources (from Legras et al. [32]). The tree was constructed from the chord distance between strains based on the polymorphism at 12 loci and is rooted according to the midpoint method. Branches are coloured according to the substrate from which strains have been isolated. The percentage of occurrence of nodes obtained through a Jacknife procedure is given at the basic node of main groups (blue figures). Type strain CBS1171 is given in black. Strains closely related to 522 Davies, or to the Champagne strain EC1118, are indicated by “522 Davies” and “Champagne group”.
Color code: Wine dark green; Cider light green; Bread yellow; Beer orange; fermented milk pink; Sake from Japan dark blue; Chinese, Rice wine and distillery from Vietnam and Thailand light blue; sorghum beer or palm wine from Africa brown; Oak tree from America blue-green; distillery from South America and rum from French Indies purple; Laboratory strains red.
Misc.: Miscellaneous
The scale corresponds to a Dc chord distance of 0.1.
For wine yeasts, 95% of strains isolated around the world belong to the same cluster [32], suggesting a unique origin of wine yeasts, followed by expansion of populations through human activities. The initial domestication event may have been located in Mesopotamia [32]. From this area, at least two yeast migration routes can be drawn in Europe. First, from an area close to Lebanon, migration may have occurred through the Mediterranean Sea, towards Italy, Spain and France, and in France along the Rhone Valley towards Burgundy, then Alsace and from the estuary of the Loire river. This transfer of yeast strains may have been related at least partly to the transfer of the grape varieties: Ugni blanc, which originated from Italy, is now the main grape variety used in Cognac, and the Muscadet grape variety (Muscadet wine) is known to have been imported from Burgundy to Nantes during the 15th Century AD. Second, a migration route along the Danube Valley can be assessed. Probably this could also have happened via a co-migration of vine and yeast as the Traminer grape variety is found all along the Danube river valley [39].
Two attempts have been made to estimate the divergence time of the different European colonization roads. One is based on the genetic differences between the most distant wine strains [29], the other between the wine populations from Lebanon and Central Europe [32]. Both estimations agree with a divergence occurring less than 30,000 years ago. The divergence time estimation of about 11,000 years BP [32] between a Lebanese group of strains and a group of Central Europe suggests a rather early divergence, but posterior to the last glaciation era. It is however not possible to conclude whether this divergence is connected to a post-glaciation colonization route of wild vine [40] or to the culture of vine. Additional population genetics studies of yeast coming from Caucasia, the area of grape origin, as well as from Central Europe, would shed light on the origin and roads of wine making.
5 Beer and bread fermentations: interspecies hybridization and polyploids
The emergence of domesticated forms has been shown to have been driven by natural and human selection, migration, mutation, as well as genetic drift due to population bottlenecks. It requires genetic innovation in domesticated forms as well as restriction of gene flow between domesticated and natural forms. In addition to mutations, intra and inter-specific hybridization has been especially efficient for creating domesticated forms. By changing chromosome number and/or causing chromosomal imbalance, hybridization often leads to partial or complete reproductive isolation. Polyploids, aneuploids and diploid interspecific hybrids have been encountered several times in domesticated Saccharomyces sp. isolates. It is indeed now well accepted that inter-species hybridization and genome duplication have participated widely to the evolution of domesticated yeasts.
The most famous interspecific hybridization events are probably the one involved in yeast used for making lager beer. As previously described, there are two groups of beer. Ale-type beer, which is fermented using S. cerevisiae at 15–24 °C, has been brewed since ancient times, probably as early as 6000 BC [18]. In contrast, lager beer which is made via low temperature fermentation (5–14 °C) originated more recently during the Middle Ages. It is thought to be concomitant with a Bavarian law that prohibited beer fermentation in summer and therefore encouraged brewing in wintertime. Then, lager beers gained popularity in the late 1800s, concomitant with the advent of refrigeration that allows cool fermentation even in the summer. The lager yeasts commonly known under the name S. pastorianus have arisen from inter-specific hybridization and selection for low temperature fermentation. Numerous genetic studies carried out for more than two decades have proposed that the S. pastorianus genome contains different combinations of the genomes of the following closely related species: S. cerevisiae, S. bayanus and S. uvarum, and possibly a fourth unknown lager type species (for reviews, see [18,41]). Indeed, the species S. uvarum and S. bayanus are known to be able to tolerate cooler temperatures than S. cerevisiae. The presence of one of these two species among ancestors of S. pastorianus would explain the cold temperature fermentation ability of the lager yeast species. More recently, whole genome sequencing data revealed that S. uvarum was not an ancestor of lager yeast as previously thought but rather that lager beer yeasts were hybrids between S. cerevisiae and S. bayanus-like strains with possible other unknown lager type species [33,41]. Interestingly, the extensive analysis of the genome organization of numerous S. pastorianus strains coming from various geographical locations and beer types have revealed that at least two hybridization events have occurred in lager beer [41,42]. The first one has led to the yeast used for making beer known as Saaz-type (usually brewed in the Czech Republic as well as in Calsberg breweries in Denmark). The second one has led to the yeast used for making beer known as Frohberg-type (usually made in the Netherlands and in non-Carlsberg Denmark breweries). Yeasts of Saaz-type lager beer have a diploid DNA content with around an equal volume of S. cerevisiae and S. bayanus genomes. In contrast, yeast of the non Saaz-type have an equivalent of triploid DNA content with two genomes coming from S. cerevisiae and one coming from S. bayanus. The two groups of lager yeasts have S. bayanus-like mitochondrial DNA sequences but no S. cerevisiae mitochondrial DNA [41,43]. Phylogenetic tree across a large number of S. cerevisiae strains coming from different fermentation processes and other ecological niches (laboratories, forest, fruits…) suggest that the S. cerevisiae ancestor(s) of lager yeasts came from ale-type beer [41]. Other beer strains are inter-specific hybrids between S. cerevisiae and S. kudriavzevii [44–46]. In addition, interspecific hybrids between S. cerevisiae and S. bayanus have been detected in other fermentation processes such as those involving wine and cider [28,47].
From a single species, genome-doubling leading to variation of ploidy may lead to the emergence of a new species. Evidence of whole genome duplication, chromosome copy number variation or gene copy number variation has been detected several times among strains described as belonging to the S. cerevisiae species [32,48]. In domesticated S. cerevisiae strains, several strains, essentially from bakery and brewery origins, exhibited four alleles at several loci, suggesting a partial (aneuploidization) or a whole-genome duplication (polyploidization) during the domestication of S. cerevisiae yeast [31,32]. Extensive characterization of these strains indicates that most of the strains from bakery origins are indeed autotetraploids. They have originated from the hybridization of at least two S. cerevisiae strains and they have four copies of each chromosome allowing correct pairing, formation of tetravalents and segregation at meiosis. Although dating the hybridization event(s) is difficult, the tetraploidization event(s) probably occurred recently since autotetraploids with tetrasomic segregation usually evolve towards disomy quickly.
The frequency of interspecific hybridizations in the Saccharomyces sensu stricto species as well as the frequency of ploidy variation events are not known precisely but seem to be high. Usually these events are detected in domesticated yeasts suggesting that they are associated with the action of humans. Given the high frequency of these events, the definition of yeast species in the Saccharomyces sensu stricto complex becomes a subtle question. It makes however this group of species an interesting model for studying speciation processes and the role of humans in driving speciation and radiation.
6 Gene changes associated with the human exploitation of yeast
For wine yeasts, specific alleles of two genes involved in essential functions in wine making have been selected for. The first example of adaptation directly related to wine-making technology has been described by Perez Ortin and colleagues [49] about the resistance to sulfite. This antiseptic was used to clean wine containers, first by the Egyptians, later by the Greeks and Romans, and became widely used starting the Middle Ages until today [49]. These authors have characterized a translocation between chromosome VIII and XVI in the promoter region of the SSU1 gene which encodes a plasma membrane sulfite pump involved in sulfite metabolism and detoxication. This mutation provokes the induction of the SSU1 transporter and increases the ability of yeast cells to expulse sulfite from the cytoplasm and provides them for a better resistance (Fig. 4). This mutation can be detected in 50% of the wine strains [50] whereas it has not been detected among wild strains. In addition, a McDonald–Kreitman test of neutrality suggested that this locus has not evolved neutrally in a wine yeast population owing to a significant excess of replacement polymorphisms relative to the neutral expectation (P = 0.037) [51,52].
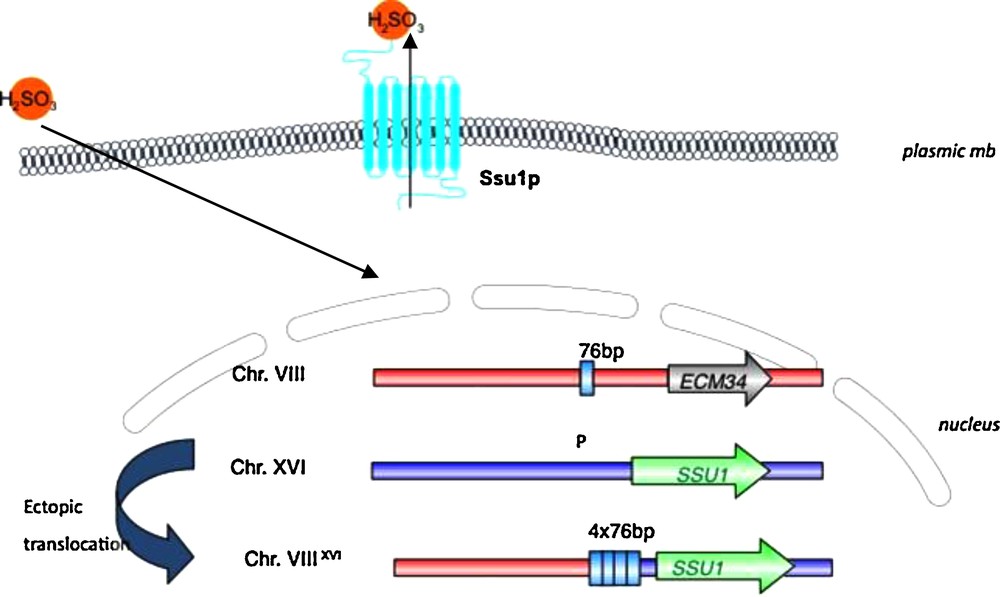
Schematic representation of Chromosome VIII – Chromosome XVI ectopic translocation that leads to an increase of gene SSU1 expression. This gene codes for a plasma membrane sulfite pump and enables yeast cells to resist to sulfite. During the translocation, a 76 bp region is placed in the promoter region of SSU1, and duplicated. The number of repeats is related to the resistance of yeast cells. “P” corresponds to the location of the promoter of SSU1.
A second example of human driven adaptation has been identified for the very specific wines of the Jerez area. These wines are submitted to a special aerobic aging process led by “Flor yeast”, a special group of Saccharomyces cerevisiae strains [53]. Flor yeast cells develop at the surface of the wine (velum), a specific ability that has been linked to the parietal protein Flo11p [54]. These cells aggregate and are able to entrap CO2 bubbles, thereby providing buoyancy [55]. Flor yeast strains have acquired two mutations in the Flo11 gene [56]. The first mutation is a deletion in the promoter which relieves the gene from repression and the second mutation leads to an increase of the protein central domain and confers a higher hydrophobicity to the cell, increasing their ability to adhere to each other. These mutations are detected in a high proportion of Jura flor strains (Legras, unpublished data).
Beer strains also developed specific mutations enabling a better adaptation to malt brewing. Indeed, the analysis of the genome of lager yeast strains revealed an increased copy number of genes involved in maltose utilization [41], the main carbon source of malt. The recent sequencing of yeast genomes has revealed the presence of new regions, containing genes whose functions are under investigation. For example, in wine yeast, one of these genes has been recently characterized as a high affinity fructose transporter [57,58], a function advantageous at the end of wine fermentation. We can expect that the coming sequencing of many wine strains will enable genomic population studies and lead to the detection of more genes under selection.
In contrast to plants, programs of yeast breeding for improvement of physiological properties have been only recently developed. Mapping of quantitative traits loci, classical introgression programs as well as marker assisted selection have been used in the last decades and will further shape domesticated yeast diversity in the future.
7 Concluding remarks
In conclusion, domestication has led to the evolution of new yeast forms and species via hybridization, polyploidization, gene duplication, and gene transfer. New physiological properties, such as sulfite resistance, have arisen through human domestication. However, except for wine yeasts, the evolutionary history and the life cycles of natural and domesticated populations remain largely unknown. The exponential growth of available genome sequences as well as the increased number of research studies on natural and domesticated populations of yeast will in a near future give new insights into the effects of human and natural selection and genetic drift, as well as on the extent of gene flow between wild and domesticated environments.
Conflict of interest statement
The authors have no conflict of interest.
Acknowledgements
We thank Aymé Spor, Olivier Martin and Jean-Claude Mounolou for critical reading of the manuscript and English language improvements.


