1 Introduction
Plant and animal domestication and their subsequent worldwide diffusion have had profound consequences on human history. It has created the ground for the rise of civilization, through the facilitation of population expansion, the explosion of technology that has accompanied agricultural development and the subsequent social stratification [1]. Tracing back the history of cultigens, from their initial domestication to their most recent improvement, has therefore always fascinated scientists starting with Darwin [2]. Understanding the evolution of cultivated species requires investigations from multiple areas of research, including archaeology, history, social sciences, botany, molecular biology and evolutionary genetics. Often, discoveries in one research area drive future investigations in another and ultimately, only the combination of all sources of data can afford a comprehensive view of domesticated species that have built the foundation of human modern societies.
Along with rice and wheat, maize is one of the world leading crops and a critical source of food, feed, fuel and fibers. As such, it has traditionally been used as a model for plant geneticists. Maize is unique in at least two respects. First, it has the broadest cultivated range of all crops, from the south of Chile 40°S to Canada 50°N, from the Andean mountains where it can grow at altitudes of 3400 m above the sea level to Caribbean islands. Second, it has an unprecedented level of morphological, nucleotide [3,4] and structural diversity [5]. Hence, two maize inbred lines have on average a greater divergence than two hominids separated by 3.5 million years [6]. While the reasons for this extraordinary diversity are still unclear, it provides a unique opportunity to assess maize genetic structure and infer hypothetical routes of its worldwide dissemination. Thereafter, we review the current knowledge on maize domestication and diffusion in the Americas that conciliates archaeological, historical and genetic data. We then focus on its recent introduction to Europe followed by subsequent breeding, which includes the development of European populations, hybrids and management of genetic resources. Finally, we discuss the genetic determinants of its adaptation and provide some guidelines for future prospects.
2 Maize domestication
The most recent maize archaeological discovery [7,8] attests its presence in the Balsas region valley of Mexico, 8700 cal. years BP (calibrated years before the present) (Fig. 1). This estimate corroborates previous genetic data establishing that maize was domesticated once in that region from the subspecies Zea mays ssp. parviglumis ∼9000 years ago [9]. Cultivated maize exhibits a profound morphological differentiation with its wild progenitor, which includes modification in vegetative architecture (i.e. reduction in branching), ear morphology (i.e. the number, position along vegetative axes, size, shape and number of rows per ear), kernel morphology (i.e. kernel shape, size, hardness) and kernel characteristics (i.e. dormancy, shattering, starch and protein content). So far, two domestication genes have been cloned: tb1, teosinte branched 1, a transcription factor whose overexpression represses the growth of axillary meristems and results in the unbranched plant architecture typical of maize [10,11]; and tga1, teosinte glume architecture 1, a transcription factor responsible for the ‘naked’ grain phenotype of maize and contributing to the organization of an ear cob being able to bear several hundreds of kernels [12].
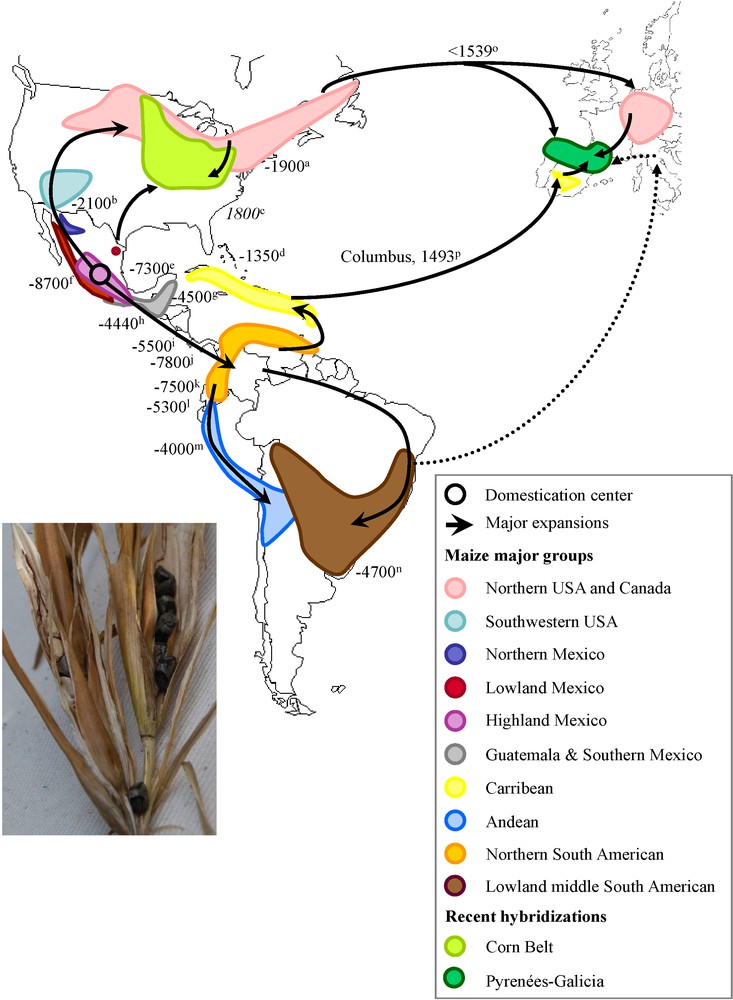
Domestication center and hypothetical diffusion of maize through the Americas and Europe. Picture on the left represents typical ears and kernels of teosinte, the wild progenitor of maize. The main maize genetic groups are represented according to [34]. Black arrows indicate the expansion scenario of maize out of the domestication area as proposed by [34], [36] and [50]. Doted arrow indicates introduction in Italy hypothesized by [50], which deserves further investigations. Negative dates are indicated in calibrated years before present and positive dates are indicated in AD. Recent hybridizations include Corn Belt dents and European maize from Pyrenees-Galicia. a: [64]; b: [65]; c: [52]; d: [66]; e: [67]; f: [7]; g: [68]; h: [69]; i: [70]; j: [71]; k: [72]; l: [73]; m: [74]; n: [75]; o: [50]; p: [44];. Masquer
Domestication center and hypothetical diffusion of maize through the Americas and Europe. Picture on the left represents typical ears and kernels of teosinte, the wild progenitor of maize. The main maize genetic groups are represented according to [34]. Black ... Lire la suite
Although the timing of maize domestication is still subject to debate, several lines of evidence suggest that this process could have been relatively fast. First, morphological observation of cobs dating back 6250 cal BP [13] and 5500 cal BP [14] from Guila Naquitz (Oaxaca, Mexico) and Tehuacan valley (Puebla, Mexico), respectively, reported the presence of non-disarticulating rachis, and exposed grains [15]; good evidence of the latter is actually already present 8700 cal BP [7]. These results have been confirmed by the genetic analysis of cob specimens dating back 4400 BP [16] showing that the cultivated alleles at three genes including tb1 and two genes influencing protein and starch quality and quantity (pbf and su1) are present. Altogether, these observations suggest first that soon after maize was brought in cultivation by early farmers in the Americas, it was already under selection for desirable traits. Second, selection may have been facilitated by the initial presence of cultivated alleles in the initial teosinte gene pool. Hence, the frequency of the Tb1 cultivated allele is > 30% in teosinte [16] and Weber et al. [17] have shown that for many of the candidate genes possibly selected during maize domestication, the allele conferring a more maize-like phenotype was always found in teosintes (sometimes at elevated frequency).
While selection at targeted domestication genes has created a local depletion of diversity [11,12,18], indirect genome wide effect of selection on genetic diversity is very limited [19], consistent with the rapid breakdown of linkage disequilibrium with distance in maize [3,20]. The genome wide reduction of diversity in maize as compared to teosintes, which is ∼20% [21], is therefore mainly attributable to a domestication bottleneck of moderate intensity [3,21,22]. Recurrent gene flow with teosintes, in particular from the subspecies mexicana to maize, may also have contributed to enhance maize diversity after its initial domestication [9]. Indeed, mexicana grows as a weed in maize fields particularly in highlands of central and northern Mexico, and persistent hybrid swarms suggest that mexicana may have contributed significantly to maize diversity [23]. This hypothesis is consistent with evidence of recent gene flow from mexicana to maize [24–26].
3 Maize diffusion through the Americas
The history of maize diffusion through the Americas from its center of origin in Mexican highlands (Fig. 1) has been assessed using different types of markers following the development of new technologies. Initial work was carried out using chromosomal heterochromatic bands (knobs) morphology and suggested that maize first spread into South America through the Andes and more recently along the east coast of Brazil [27]. Isozymes were subsequently used and established a clear distinction between maize growing at low and high elevation both in Mexico [28] and Guatemala [29]. In addition, these studies provided new highlights into the migration paths of maize, northwards from Mexico to USA and Canada [30] and southwards from Mexico to the Caribbean coast of South America to the Caribbeans, and from Mexican highlands to Guatemalan highlands (reviewed in [31]). These early studies therefore suggested the existence of two southwards routes, a highland route emerging from the Mexican highlands and reaching the Andes and a lowland route from the Mexican lowlands expanding toward the East coast of South America, middle South American maize (Bolivia, Argentina, Paraguay and Uruguay) being the meeting point of these two expansions [32].
Further examination of microsatellite variability in ancient DNA specimens supports these observations [33]. However broader microsatellite surveys [9,34] suggest instead that maize southward expansion has occurred through the lowland tropics of Mexico from Central America to Guatemala to the Columbian lowlands and subsequently followed two paths, a highland path along the Andes and a lowland path spreading in the East coast of South America and reaching the middle South America. Under this scenario adaptation to altitude would have been acquired de novo in the Andes [34]. In fact, using microsatellites, Vigouroux et al. [34] found a very poor correlation (r2 = 0.044) between genetic distance and altitude, which therefore does not account for distinction between maize growing at high and low elevations both in Mexico and Guatemala.
Beside this discrepancy, most observations from microsatellite markers corroborate earlier studies [9,34–37] that:
- • Caribbean maize was imported from South America from Trinidad and Tobago;
- • lowland South America is the contact zone between the highland path (Andes) and the lowland path (East Coast of South America);
- • northward expansion has occurred through northern Mexico, south western USA to northern USA and Canada;
- • southeast USA material (corn belt mid western dents) is a hybrid derivative from southern dent-northern flint crosses. Maize cultivation in northern regions is attested between AD (Anno Domini) 800 and 1100 [38], when maize rapidly replaced sunflower. It also formed the staple food for the mixed agricultural hunter gatherers Iroquoian populations that populated North America up to the Saint Laurent at the time of Discovery [39].
Today the structure of maize genetic diversity classifies all races in four genetic clusters that are poorly differentiated [34]:
- • the Mexican highlands that cover highland Mexico, northern Mexico and south western USA and most lowland Mexico;
- • the tropical lowlands that cover Guatemala and southern Mexico, the Caribbeans, northern South American and middle South American maize;
- • the Andean group encompassing the Andean maize and part of the middle South American;
- • the northern USA group that includes northern USA and Canada as well as some south Andean maize or middle American maize that are not shown on the map in Fig. 1 but were imported form northern USA recently [34,36]. Note that this last group appears particularly divergent from any other maize race [37], although it is hypothesized to have been derived from southwestern USA material. Interestingly, among the 310 races analyzed in [34] 44% have an ancestry in more than one of the clusters, underlying the complex history of maize that has been punctuated by germplasm exchange, admixture and potential long distance gene flow [40,41].
4 Maize introduction to Europe and subsequent utilization
The use of local names for maize in Europe, such as “Turkish Corn” in Germany, was first taken as an evidence for its introduction from Middle East (discussed in [42]). But its American origin has since been well established by the historian Matthioli [43]. The first historical record attesting maize European introduction is that from the Caribbean by Colombus in Spain, 1493. From there, maize was rapidly brought to the Vatican [44] and the earliest European representations of maize are most likely those painted in frescoes near Rome dating from 1517 [42]. While Caribbean tropical maize was likely not adapted to cultivation beyond the warm regions of southern Europe (see paragraph 5, [44]), its cultivation in northern European regions has been unambiguously reported since 1539 in Germany by the German herbalist Bock [45], ‘in all gardens, almost everywhere’ according to [46]. Maize was also already well established in Alpine regions of Italy by 1570 [43,47]. These results suggest either a rapid expansion from southern Europe accompanied by adaptation to low temperatures, or the existence of a second distinct introduction of maize pre-adapted to temperate climates. Interestingly, early botanical records describe two highly distinct morphotypes [45] from the Americas. Maize became popular during the 17th century in northern Spain [48] and southwestern France [49].
Large-scale molecular analyses of maize from both Europe and the Americas [36,50] have confirmed that the diversity of European maize derives from that of the American maize and represents ∼75% of the latter, illustrating the bottleneck inherent to all introductions. These results have also revealed two important observations: (i) several accessions cultivated in southern Spain are related to Caribbean maize, consistent with an introduction by Colombus; and (ii) traditional landraces from northern Europe are instead related to North American northern flint varieties. The latter supports an independent introduction of northern American materials pre-adapted to northern European climate (Fig. 1), soon after Discovery as first hypothesized by [45]. History of the Discovery of North America [51] has revealed numerous expeditions between 1492 and 1539, both official expeditions from England, Spain and France (often involving multinational crews), and unofficial fishing expeditions toward the Saint Laurent, thereby offering numerous opportunities for a second maize introduction to Europe (see [36,50]) at a time consistent with the broad scale cultivation in Germany reported by [45] and [46]. Subsequent introductions from northern America have probably occurred as documented in 1604 for northern Spain [48]. In addition similarities between Italian traditional varieties and a few varieties from South America, suggest the contribution of a third introduction (dotted arrows in Fig. 1).
Interestingly, molecular analyses have also revealed that European varieties cultivated at mid-European latitudes (from the Pyrenées mountains, to northern Portugal) have no direct American counterpart but share alleles from both northern flints and tropical lowlands. Northern flint material therefore appears to have contributed to adaptation in these regions, similar to what has been documented in the Corn Belt, where farmers crossed “southern dents” from tropical origins with northern flints to adjust flowering time [52,53]. Independent introductions, hybridization and local selection have shaped new genetic groups specific to European maize [54]. Hence, mid-latitude European materials are now sufficiently divergent from founder gene pools to appear as distinct genetic groups rather than hybrids in clustering analyses [54].
After World War 2, European traditional varieties were used to develop hybrid varieties [55] adapted to northern European conditions. Hence, inbred lines derived from the French southwestern Lacaune population by André Cauderon and co-workers, namely F2 and F7 (for France n°2 and 7, respectively), were crossed to inbred lines developed by USA research. The resulting hybrids (Fig. 2) over yielded by 20–25% the varieties imported from the USA [56] and extended maize cultivation North of the Loire Valley. The use of genetic complementarities between the highly divergent European flints and the USA dents still forms the core of most hybrid breeding programs in northern Europe. Hybrid varieties later outperformed and replaced local open pollinated varieties from southern western France, Bresse and Alsace. Genetic studies illustrate that the evolution from traditional varieties to hybrids and further hybrid breeding has led to a shift in the allelic composition of the cultivated gene pool without any severe depletion of diversity [57]. Nevertheless, European traditional varieties are being preserved in collections and hence accessible for breeding purposes.

F2 and F7 are two European flint lines that were crossed with a US dent line (W182E) to form the French variety INRA 260. The INRA 260 hybrid was released in 1960 and was largely cultivated in France in the 1960s (source A. Cauderon and X. Lascols).
5 Maize adaptation
Maize expansion has been accompanied by a dramatic adaptation to environmental conditions, in particular the adjustment of plant cycle to the duration of the growing season. Gouesnard et al. [58] have illustrated that tropical maize accessions display a very large variability for duration of planting to flowering both in their original short day environment and under long days, with substantial variation in photoperiod sensitivity. However, none of these materials are able to produce grains in northern Europe. The initial range of variation has therefore been expended by the selection of early flowering types adapted both to a short growing season and low temperature.
Large scale QTL mapping from multiple crosses has revealed that variation in flowering time is controlled by at least 50–60 loci (QTL) with mild effects [59,60]. These loci affect flowering time through either sensitivity to photoperiod [61] or intrinsic earliness. Most of them display complex allelic series with at least three classes of effects [59]. Understanding the nature of allelic series at these QTLs is now possible thanks to association mapping [20] and linkage-based fine mapping [62]. The most advanced case study is the locus vgt1 cloned by Salvi et al. [62], which regularly appeared in QTL mapping studies and was confirmed to display a strong effect on flowering time by association mapping [63]. Vgt1 corresponds to a 2.7 kb non coding region located upstream of gene ZmAp2, and controls its expression. Polymorphism at vgt1 is structured into major haplotypes that can be discriminated by the absence or presence of a short transposable element (a MITE, Miniature Inverted repeat Transposable Element). The absence or presence of the MITE marks the early and late flowering alleles, respectively. Those alleles are characterized by a seven day difference in flowering time in climatic conditions of northern France. Frequency of alleles at vgt1 among a set of 256 populations (Fig. 3) reveals that (i) both alleles have coexisted with balanced frequencies in Mexican highlands, where farmers manage maize varieties with contrasted cycle duration, (ii) the early allele was eliminated in lowland warm environments where early flowering is deleterious because of its negative impact on plant size and yield, and (iii) the early allele was selected positively during migration in cold temperate environments, i.e. along the “northern flint migration route”, thereby contributing to its rapid success in northern Europe. Note that although earliness has been favored in the high elevation of Andes, the early allele at Vgt1 is not found in this material, consistent with the tropical lowland origin of this group.
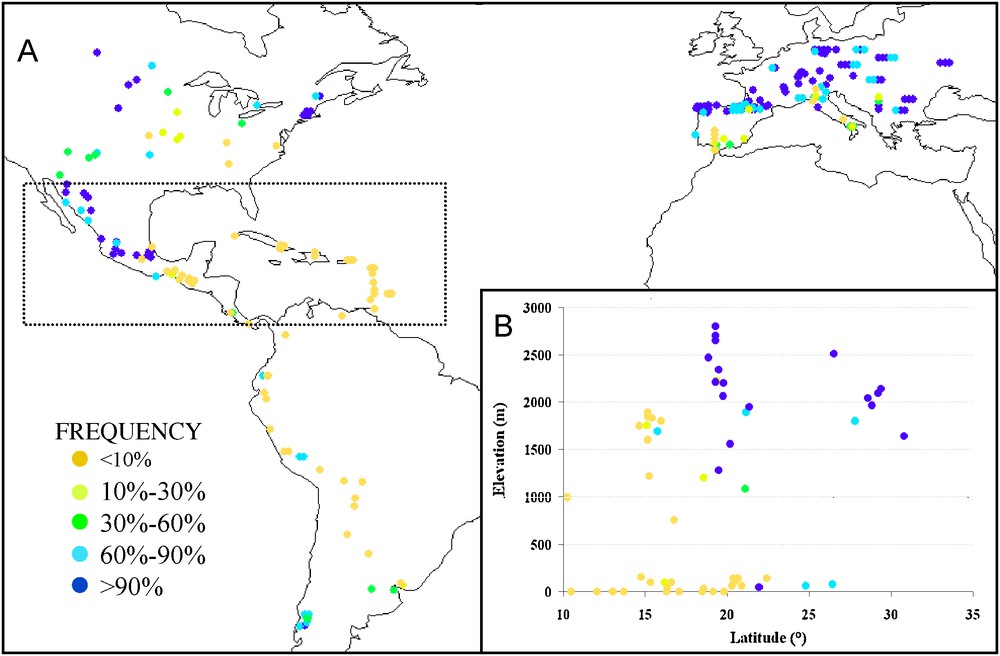
A: Geographical distribution of the MITE insertion associated with early flowering at the Vgt1 locus in 256 European and American landraces. B: MITE frequency along latitude (X-axis) and elevation (Y-axis) for a subset of 77 landraces from Central America and the Caribbean (delimited by the rectangle in A).
6 Conclusion
Maize is certainly one of the most striking examples of plant domestication, with two genes conferring radical changes in plant architecture and grain type, thus explaining its wide distribution and success in agriculture. Recurrent gene flow with the original wild gene pool has contributed to retain a large fraction of the genetic variability of the wild gene pool, facilitating maize extreme diversification. From an original pool of tropical varieties un-adapted to cultivation under the long days and low temperatures of temperate climates, Amerindian populations and, later, farmers and breeders have successfully selected varieties adapted to cultivation up to 50° North. This was accompanied by a strong selection at target genes and a complete reorganization of the genetic diversity through selection, inbreeding and hybridization. Efforts are now devoted to the conservation of this genetic patrimony, which presently receives increasing attention for direct cultivation, for cultural demonstration and/or alternative farming systems, and for genetic studies aiming at identifying genes and alleles of interest. Such alleles will contribute to sustain breeding efficiency thereby meeting the new challenges raised by the evolution of mankind.
Conflict of interest statement
The authors have no conflict of interest.
Acknowledgements
Alain Charcosset is grateful to Pierre Dubreuil, Cécile Rebourg, B. Gouesnard, M. Chastanet and Sébastien Ducrocq for their contribution to the results presented here and to Céline Mir for ongoing research.



Vous devez vous connecter pour continuer.
S'authentifier