1 Introduction
The Northern Pike Esox lucius Linnaeus 1758 (Teleostei, Esocidae) is globally the most common esocid (although this might differ locally). Its distribution is circumpolar, covering both North America and Eurasia [1]. It holds high socio-economic interest for recreational and commercial fishing [1,2]. Moreover, pike reproduction is well known [3]. Aquaculture is now well-developed and has been used to restock numerous waterbodies [1–4] or to introduce it in places where it is not native [5,6].
Since the early 19th century, the taxonomy of the Esocidae is considered to be well known, and E. lucius was thought to be the only species present in Europe [1,7]. Faced with the high ecological variability of pikes, Nilsson et al. [8] asked “How many species of pike are there?”. Yet neither they nor other researchers thought that many new species of pikes remained to be described, especially in Europe where the vertebrate fauna was assumed to be well known. Numerous genetic studies proved low variability between American and North European Northern Pike populations [9–12]. A contrario, however, South-European populations present a higher variability, especially in Adriatic drainages and Italy [13,14], in the Danube basin and the Southwest of France [15]. However, during the last decade, other groups of European freshwater fishes turned out to include many cryptic or just undetected species [16,17].
Very recently, Bianco and Delmastro [18] and Lucentini et al. [6] described, independently, the same endemic species of pike in Italy, and named it, respectively, E. cisalpinus and E. flaviae, the first being the valid name according to the priority rule of the International Code of Zoological Nomenclature, E. flaviae being a junior synonym [19]. This species differs from E. lucius by having 92–107 lateral scales (vs. 105–148), 4–4 submandibular pores (vs. 5–5), a banded or reticulated pigmentation on the sides (vs. rounded spots) [18], and an average divergence of 1.80% on the cytochrome c oxidase subunit 1 mitochondrial marker (COI) [16]. This genetic divergence is also supported by microsatellite markers [13,14]. This species is present in the Padany-Venetian and Tuscany-Latium districts in Italy, but was extensively bred and introduced elsewhere in Italy as well as in other countries such as France during the last 50 years [19].
In France, pike, considered by default to be E. lucius [20], is native to the Rhine, Seine, Loire (except in Brittany), and Rhône drainages. There are records of its introduction in the small coastal French Mediterranean rivers out of the Rhône itself [21]. Archeological data indicate its presence in Aquitania during the Pleistocene [22], suggesting that it is native to the Dordogne and Garonne rivers and maybe also in the Adour drainage.
Launey et al. [15] used a microsatellite approach on several pike populations from France and the East of Europe to assess the impact of pike restocking on the genetic diversity of native populations. They found extensive introgression between wild populations and introduced stocks, but interestingly also highlighted a very divergent population from the Southwest of France, very different from all their other French and Eastern European samples. Chimits [23] had already pinpointed an earlier spawning period in this region compared to other French pike populations.
The aim of our study is to review the taxonomy of pikes present in France using an integrative taxonomy approach [24]. We use morphological and molecular data on 19th century and recent specimens collected in the main catchments of France to explore the diversity of French pikes. We replace them in the context of the variability of European pikes (including the description of an eventual new species), and investigate the presence of E. cisalpinus in France using the characters of the two descriptions [6,18].
2 Materials and methods
2.1 Sampling and measurements
The “Muséum national d’histoire naturelle” (MNHN, Paris) and the Claude-Bernard – Lyon-1 University, with the collaboration of the “Office national de l’eau et des milieux aquatiques” (ONEMA), of the “Fédération nationale de la pêche en France” (FNPF), and of some professional and amateur fishermen, performed sampling operations from 2003 to 2013. Sixty-five specimens were caught by electrofishing or angling in 32 locations from the main French drainages (Seine, Loire, Garonne, Rhine, Rhône, Adour) (Table 1). Samples were collected in rivers classified as ‘French first category’ (cold salmonid rivers where pike populations are supposed to be native because its introduction is strictly prohibited [15]), and in rivers classified as “second category”, i.e. warmer cyprinid rivers with mainly limnophylic species. In order to have a link with the study of Launey et al. [15], we collected in the exact location (Boutonne stream) where the very divergent population was noticed, and one specimen could be captured. Because pike has a vulnerable protection status in France, most specimens were released alive after having been photographed and sampled for molecular study. Some specimens were kept as vouchers. They were fixed in formalin (or ethanol for the smallest ones). They were deposited in the collections of the MNHN or of the Claude-Bernard – Lyon-1 University and were used for the morpho-meristic study.
Sampling sites and GenBank accession numbers (here provisional numbers) for COI and Plagl2 sequences of the French individuals.
| Drainage | Location | Sample ID | (provisional) GenBank accession numbers | |
| COI | Plagl2 | |||
| Adour | Adour at Estirac | BRO462 | BRO462 | BRO462 |
| Estampon at Saint-Gor | BRO531 | BRO531 | BRO531 | |
| Geloux at Garein | BRO534 | BRO534 | BRO534 | |
| Charente | Antenne at Le Seure | EM17879 | EM17879 | – |
| EM17880 | EM17880 | EM17880 | ||
| Boutonne at Saint-Séverin-sur-Boutonne | BRO545 | BRO545 | BRO545 | |
| Charente at Saint-Saviol | BRO25 | BRO25 | BRO25 | |
| Lien at Condac | BRO505 | BRO505 | BRO505 | |
| BRO506 | BRO506 | BRO506 | ||
| BRO509 | BRO509 | BRO509 | ||
| Seugne at Les Gonds | BRO29 | BRO29 | BRO29 | |
| Sonsonnette at Saint Front | BRO433 | BRO433 | BRO433 | |
| Dordogne | Dordogne at Cénac-et-Saint-Julien | BRO19 | BRO19 | BRO19 |
| BRO22 | BRO22 | BRO22 | ||
| Isle at St-Médard-de-Guizière | FFFtag12251 | FFFtag12251 | FFFtag12251 | |
| BRO453 | BRO453 | BRO453 | ||
| BRO455 | BRO455 | BRO455 | ||
| BRO457 | BRO457 | BRO457 | ||
| Isle at Trélissac | BRO24 | BRO24 | BRO24 | |
| Lary at St-Martin-d’Ary | EM17877 | EM17877 | – | |
| Eyre | Eyre at Bélin-Béliet | BRO441 | BRO441 | – |
| BRO443 | BRO443 | BRO443 | ||
| BRO445 | BRO445 | BRO445 | ||
| BRO536 | BRO536 | BRO536 | ||
| Eyre at Mios | BRO21 | BRO21 | BRO21 | |
| BRO23 | BRO23 | BRO23 | ||
| Grande Leyre at Sabres | BRO538 | BRO538 | BRO538 | |
| BRO541 | BRO541 | BRO541 | ||
| Garonne | Garonne at Verdun-sur-Garonne | BRO20 | BRO20 | BRO20 |
| Loire | Boivre at Béruges | BRO502 | BRO502 | BRO502 |
| Sèvre Nantaise at Saint-Malo-du-Bois | BRO1 | BRO1 | BRO1 | |
| Meuse | Meuse at Han s/Meuse | BRO6 | BRO6 | BRO6 |
| BRO7 | BRO7 | BRO7 | ||
| BRO8 | BRO8 | BRO8 | ||
| Rhône | Clauge at La Loye | BRO427 | BRO427 | BRO427 |
| BRO428 | BRO428 | BRO428 | ||
| BRO430 | BRO430 | BRO430 | ||
| BRO431 | BRO431 | – | ||
| Chautagne at Lône-du-Brotalet | BRO14 | BRO14 | BRO14 | |
| Lake Bourget | BRO3 | BRO3 | BRO3 | |
| BRO4 | BRO4 | BRO4 | ||
| BRO5 | BRO5 | BRO5 | ||
| Rhône at Breignier | BRO13 | BRO13 | BRO13 | |
| BRO26 | BRO26 | BRO26 | ||
| BRO27 | BRO27 | BRO27 | ||
| BRO28 | BRO28 | BRO28 | ||
| Rhône at Massigneu de Rives | BRO529 | BRO529 | BRO529 | |
| Rhône at Saint-Vulbas | BRO12 | BRO12 | BRO12 | |
| Sarthe | Sazée at East of Segré | BRO15 | BRO15 | BRO15 |
| BRO16 | BRO16 | BRO16 | ||
| BRO17 | BRO17 | – | ||
| BRO18 | BRO18 | – | ||
| Seine | Blaise at Saint-Ange-et-Torçay | BRO525 | BRO525 | BRO525 |
| Epte at Guerny | FFFtag10874 | FFFtag10874 | – | |
| Serein at Pontigny | BRO464 | BRO464 | BRO464 | |
| BRO466 | BRO466 | BRO466 | ||
| BRO470 | BRO470 | BRO470 | ||
| Superbe at Pleurs | BRO9 | BRO9 | BRO9 | |
| BRO10 | BRO10 | – | ||
| BRO11 | BRO11 | – | ||
| Somme | Canal de la Maye at Favières | BRO2 | BRO2 | BRO2 |
Forty-nine specimens from the historical collections of the MNHN and the British Museum of Natural History (BMNH, London), covering the whole native distribution area of E. Lucius, were added to the morpho-meristic study [25]. The possible type of E. lucius (BMNH 1853.11.12.114) was also included in the analyses, but as only its skin on paper is conserved, no morphometric measurements were possible. We used the meristic data of the holotype of E. cisalpinus (IZA 111) from the species description. Counts and measurements were taken from the best preserved side (by default the right side). Measurements were taken with an electronic caliper and are expressed to the nearest tenth of a millimeter. For measures longer than 300 mm, we used tape measures expressed to the nearest millimeter. The standard length was measured from the tip of the snout to the basis of the uppermost caudal ray. The post-dorsal length was measured from behind the base of the last dorsal fin ray to the basis of the uppermost caudal ray. The length of the caudal peduncle was measured from behind the base of the last anal fin ray to the basis of the lowermost caudal ray. The transverse scales counts (from the pelvic fins origin to the lateral line, and from the dorsal fin origin to the lateral line) did not include the scale of the lateral line. All measurements were made point to point, never by projections. Vertebrae were counted to the ural 1 bone included. Vertebrae and unpaired fin rays were counted by X-rays (Faxitron Model 43855F, X-ray energies used: 40 to 70 kV depending on the size of specimens). When available, photos of released specimens were used for lateral scales count as additional data. In vivo observations were performed on adults and juveniles. Sex determination was realized following the criteria of Raat [1].
2.2 Molecular study
The molecular study is in two parts: a DNA taxonomy study sensu Tautz et al. [26] with recent DNA using mitochondrial (COI) and nuclear (Pleiomorphic adenoma gene-like 2 Plagl2, [27]) genes; and a DNA barcoding study sensu Hebert et al. [28] using a fragment of COI (131 bp) [29] for molecular identification of old museum collections.
All captured specimens were included in the DNA analyses. For each specimen (Table 1), a small piece of fin was stored in 95% ethanol at 3 °C. DNA extraction was performed on an EpMotion Robot using MN Biomedical extraction kits, according to the manufacturer's protocols. Twelve ancient specimens from the MNHN collections were selected based on their conservation fluid (ethanol, with no fixation in formalin). The DNA was extracted using a classical CTAB protocol with a chloroform isoamylalcohol step [30] (Table 2). For the ancient specimens, particular precautions were taken to avoid contamination, and two independent extractions and PCRs were performed to check the sequences. DNA amplification was performed by PCR in a final 20-μL volume containing 5% DMSO, 1 μL of dNTP 6.6 mM, 0.15 μL of Qiagen Taq DNA polymerase, using 2 μL of the buffer provided by the manufacturer, and 0.4 μL of each of the two primers at 10 pM; 2.5 to 10 μl of DNA extract was added. The different primers used are: for COI TelF1 5′-TCG ACT AAT CAY AAA GAY ATY GGC AC-3′, TelR1 5′-ACT TCT GGG TGN CCA AAR AAT CAR AA-3′ [31], FishR1 5′-TAG ACT TCT GGG TGG CCA AAG AAT CA-3′ [32]; Plagl2 plagl2_F9 5′-CCA CAC ACT CYC CAC AGA A-3′, plagl2_R930 5′-TTC TCA AGC AGG TAT GAG GTA GA-3′, plagl2_F51 5′-AAA AGA TGT TTC ACC GMA AAG A-3′, plagl2_R920 5′-GGT ATG AGG TAG ATC CSA GCT G-3′ [27]. A new reverse primer was developed specifically for Esox sequences to amplify a small COI fragment (131 bp) for the study on Museum DNA: EsoxminibarR130 5′-AAG ATT ATW ACR AAR GCA TGG GCT G-3′.
Samples from historical collections selected for molecular work. When two specimens shared a jar and a collection number, they were singled by size or attachment of the collection label. Specimens have been identified morphologically before analyses. BOLD sample IDs are given for samples for which the amplification of the COI short fragment was successful.
| Morphological identification | Collection number | Country | Sampling location | Collector | Remark | BOLD sample ID |
| Esox lucius | MNHN A-9974 | France | Lake Grand-Lieu (Loire) at Grand-Lieu | Thomas (1858) | – | |
| Esox lucius | MNHN B-0941 | Switzerland | Lake Zug (Rhine) | Major | – | |
| Esox lucius | MNHN B-0946 | France | Rhône at Avignon | Blanchard (1880) | – | |
| Esox nov. sp. | MNHN B-0944 | France | Charente at La Rochelle | D’Enfer (1824) | – | |
| Esox nov. sp. | MNHN B-0945 | France | Lake Mouriscot (Adour) at Biarritz | Blanchard (1880) | Smallest | MNHN B-0945a |
| With label | MNHN B-0945b | |||||
| Esox cisalpinus | MNHN B-0942 | Italy | Reno at Bologna | Savigny (1823) | – | |
| Esox cisalpinus | MNHN B-0947 | Italy | Lake Trasimeno (Tibre) | Canali | No label | MNHN B-0947 |
| With label | – | |||||
| Esox cisalpinus | MNHN B-0948 | Italy | Lake Como (Po) | Pentland | – | |
| Esox cf. cisalpinus | MNHN B-0949 | Switzerland | Lake Geneva (Rhône) at Geneva | Candolle | With label | MNHN B-0949a |
| No label | MNHN B-0949b |
After denaturation for 2 min, the PCR was run for 50 to 60 cycles of (20 s, 94 °C; 20 s, 50 °C; 50 s to 1 min 10 s, 72 °C) on a Biometra trioblock (T3000) or a Biorad Applied 2700 cycler. Successful PCRs were selected on ethidium-bromide stained agarose gels. Sanger sequencing was performed by a commercial company (Eurofins; http://www.eurofins.fr) using the same primers.
All sequences were obtained for both the reverse and forward primers. Chromatograms in both directions were compared using CodonCode Aligner 3.9 (Codon Code Corporation) and automatic base calls were checked along the sequence, both where the two sequences were in disagreement and elsewhere. The sequences were obtained (PCR and sequencing) at the same time as for species from very divergent groups, so contaminations would be more visible. Any dubious sequence (high divergence, unexpected placement in the tree) was resequenced from an independent PCR. Esox Plagl2 sequences include a relatively long microsatellite at one end that impacted sequence quality. Sequences were trimmed to retain only the high-quality part (531 bp). The Plagl2 outgroup used allowed one to identify the synapomorphies comparing it with E. lucius and E. aquitanicus sequences. This yielded one dataset for the partial COI gene (650 bp) and one for the nuclear Plagl2 marker (531 bp). We also included the sequences available in GenBank from other Esocids from all over the distribution area for both the ingroup and as outgroups (COI: E. americanus americanus Gmelin 1789: EU52476; E. americanus vermiculatus Lesueur 1846: EU52473; E. cisalpinus [labelled in GenBank as E. flaviae]: HM563688, HM563691-92, HM563694-98, HM563700, HM563703-07; E. lucius: EU524578, EU524580-83, EU524585-92, FJ890069-71, HQ600728-29, HQ960518-22, HQ960531, HQ960615, HQ960640-41, HQ960650-51, HQ960671, HQ960745-47, HQ960799, HQ960989-90, HQ960994, HQ961032-34, JQ623940, KC500713-32; E. masquinongy Mitchill 1824: EU524594; E. niger Lesueur 1818: EU524606; Plagl2: E. reichertii Dybowski 1869: JN132603). All new sequences were deposited in the Barcode of Life database (FRBRO project) [33] and GenBank with their voucher information.
Alignment was performed manually as neither marker includes indels.
A phylogenetic analysis was performed on the COI dataset using Bayesian inference (MrBayes 3.2, [34]). A model partitioned by codon position was computed by JModelTest 2.1.1 [35]. According to the results, four independent analyses with a GTR + I + G model were run for 10 million generations, sampling every 200 generations. Ten percent of the trees were discarded as burn-in, after having checked that it was sufficient for convergence. After checking convergence had been reached, the trees and parameters resulting from the four analyses were pooled and combined in a consensus. Intra- and inter-specific distances (p-distances) were calculated with software MEGA 5 [36]. The same analysis was done with the Plagl2 gene, with a HKY model, and separating heterozygous alleles. An NJ distance tree with the Kimura 2 parameter model [37] was built to perform hierarchical placement to identify the short sequences. They were also identified using blast in BOLD and GenBank, and comparison with sequences from our complete COI dataset at the diagnostic sites. The robustness of the cluster nodes was estimated by the bootstrap method [38] with 1000 replicates.
3 Results
3.1 Morphological variability among French pikes
The 49 specimens from the MNHN and BMNH collections were morphologically identified using the diagnoses of Lucentini et al. [6], Bianco and Delmastro [18], and Casselman et al. [39], using the color pattern of the coat, lateral scales count, and submandibular pores.
Six Italian individuals with 102 to 113 lateral scales, and a color patterns of diagonal bars or stellated spots [6] were re-identified as E. cisalpinus, and so was a French specimen (MNHN 2003-0242) from Lake Saint-André (between Chambéry and Grenoble) with 113 lateral scales and four submandibular pores (but without any visible color pattern because of long formalin fixation).
Two specimens (MNHN B-0949) from Lake Geneva « lac Léman » in French have 111 and 113 lateral scales, five submandibular pores, but their color pattern has been altered by ethanol. Without any other morphological characters, they were identified morphologically as E. cf. cisalpinus.
Three ancient specimens from southwestern France (Charente MNHN B-0944 and Lake Mouriscot MNHN B-0945) have 104 and 108 lateral scales. But four individuals recently caught in the same area (MNHN 2013-0838, 2013-0389, 2013-1245, 2013-1246) show a low lateral scale count too, as well as a shorter snout, and a different color pattern with a width of 1–1.5 scale tending to split into small irregular white blotches, conferring a marbled aspect. These specimens seem to belong to a distinct lineage and were provisionally identified as an unknown species Esox sp. Two individuals from the Charente drainage (Lien stream, MNHN 2013-1247) present a combination of color pattern and lateral scale counts of both species: the smallest one has a E. lucius color pattern and a low lateral scale count (112), whereas the biggest one has a higher, E. lucius characteristic, lateral scale count (117) and a divergent color pattern.
All the other specimens have white blotches, 117 to 148 lateral scales, and five submandibular pores and were identified mainly as E. lucius.
3.2 Molecular evidence of a new species of pike in the Southwest of France
The phylogenetic tree-based on the COI among 140 individuals (Fig. 1a) groups all pikes caught in France within the subgenus (Esox) [1,40].
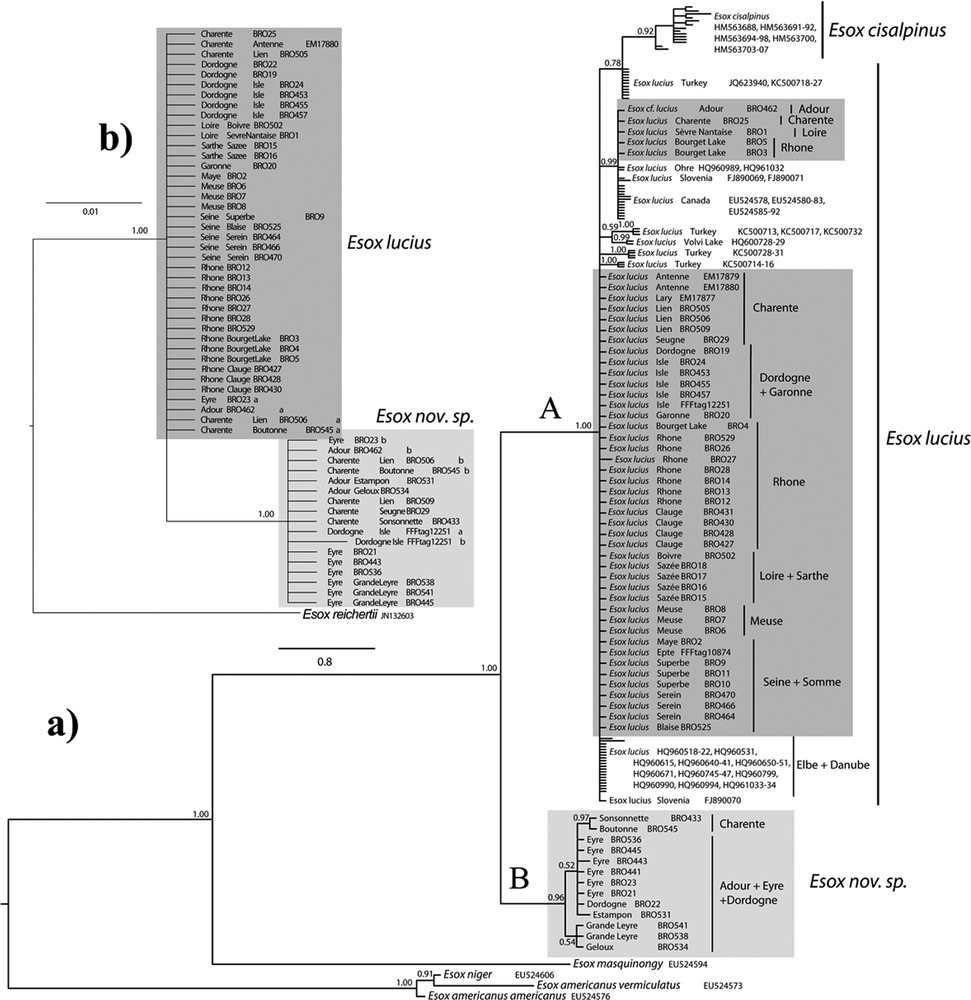
a: Bayesian tree of the cytochrome c oxidase subunit I (COI) for 140 individuals of Esox lucius and other esocids. The mean a posteriori values of the parameters are (respectively for first, second and third codon position): TL = 27.534122; alpha = 96.371958, 0.053006, 108.993368; pinvar = 0.699196, 0.944113, 0.027670; b: Bayesian tree of the pleiomorphic adenoma gene-like 2 (Plagl2) alleles for 53 French esocids. TL = 0.374231. Suffixes “a” and “b” represent both alleles from heterozygous specimen. Numbers on the nodes represent posterior probabilities. Drainage origins of the French samples are highlighted with boxes: dark grey for Esox lucius and light grey for Esox nov. sp.
Two clades are clearly separated, with a 4.0% mean pairwise divergence, with 24 diagnostic sites and high posterior probabilities. The first clade (A) is a large polytomy including samples identified as Esox cisalpinus and E. lucius. The 14 E. cisalpinus samples form a monophyletic cluster with a mean intraspecific divergence of 0.4% and 14 distinct haplotypes within a larger E. lucius cluster. The mean inter-specific divergence with E. lucius is 1.6%, and there are 11 diagnostic sites. The 109 E. lucius samples do not form a monophyletic group because of a few Turkish specimens grouped with E. cisalpinus with low support (0.78 ppv). The mean intraspecific divergence of E. lucius is 0.3% and there are 13 distinct haplotypes (three from France). There are two main groups of French haplotypes. The largest one includes samples from Charente, Dordogne, Garonne, Loire, Meuse, Rhône and Seine drainages, and their haplotypes are identical to those from the Elbe and Danube drainages. The other group includes haplotypes from the Adour, Charente, Loire and Rhône drainages, and are closed to Canadian and some Central European pikes.
The second clade (B) includes five very distinct haplotypes from southwestern French basins: Adour, Charente (including the Boutonne stream), Eyre, and Dordogne. The distance within this clade is low (mean 0.2%), and no groups are supported or organized by area within it, except for samples from the Charente drainage (0.97 ppv). This clade is very distinct from the rest of the E. lucius COI sequences covering the variability from all over its distribution area. It is sister group to the cluster formed by E. cisalpinus and E. lucius. It is also distinct and remote from all other Esox species integrated in the analysis.
The Plagl2 phylogeny of 53 individuals (Fig. 1b) shows two haplotype assemblages characterizing E. lucius and samples from this second divergent Esox clade, and diverging by three diagnostic sites (Table 3). Plagl2 identification is in agreement with the morphological and COI results. However, four specimens from Adour, Charente (including the Boutonne stream) and Eyre basins (BRO23, 462, 506, and 545) are heterozygous and have both Plagl2 alleles. These specimens are probably hybrids between the divergent and the common Esox, and some of them combine the morphological characters of both. Moreover, two specimens identified morphologically and with COI as E. lucius are homozygous for the divergent Esox Plagl2 allele, and conversely BRO22 has the COI haplotype of the divergent Esox and is homozygous for the Plagl2 haplotype of E. lucius.
Diagnostic sites and probable hybrids for the Pleiomorphic adenoma gene-like 2 (Plagl2) sequences of the 52 individuals of French pikes; one E. reichertii sample was included as outgroup.
| Identification | Sampling location | Sample IDs | COI clade affiliation | Plagl2 position | ||||||||
| 69 | 120 | 156 | 282 | 331 | 354 | 366 | 372 | 522 | ||||
| Esox lucius | Charente | BRO25 | A | A | C | G | C | A | C | T | G | A |
| Esox lucius | Antenne (Charente) | EM17880 | A | A | C | G | C | A | C | T | G | A |
| Esox lucius | Lien (Charente) | BRO505 | A | A | C | G | C | A | C | T | G | A |
| Esox cf. lucius | Dordogne | BRO22 | B | A | C | G | C | A | C | T | G | A |
| Esox lucius | Dordogne | BRO19 | A | A | C | G | C | A | C | T | G | A |
| Esox lucius | Isle (Dordogne) | BRO24, 453, 455, 457 | A | A | C | G | C | A | C | T | G | A |
| Esox lucius | Boivre (Loire) | BRO502 | A | A | C | G | C | A | C | T | G | A |
| Esox lucius | Sèvre nantaise (Loire) | BRO1 | A | A | C | G | C | A | C | T | G | A |
| Esox lucius | Sazée (Sarthe) | BRO15, 16 | A | A | C | G | C | A | C | T | G | A |
| Esox lucius | Garonne | BRO20 | A | A | C | G | C | A | C | T | G | A |
| Esox lucius | Maye (Somme) | BRO2 | A | A | C | G | C | A | C | T | G | A |
| Esox lucius | Meuse | BRO6, 7, 8 | A | A | C | G | C | A | C | T | G | A |
| Esox lucius | Superbe (Seine) | BRO9 | A | A | C | G | C | A | C | T | G | A |
| Esox lucius | Blaise (Seine) | BRO525 | A | A | C | G | C | A | C | T | G | A |
| Esox lucius | Serein (Seine) | BRO464, 466, 470 | A | A | C | G | C | A | C | T | G | A |
| Esox lucius | Rhône | BRO12, 13, 14, 26, 27, 28, 529 | A | A | C | G | C | A | C | T | G | A |
| Esox lucius | Lake Bourget (Rhône) | BRO3, 4, 5 | A | A | C | G | C | A | C | T | G | A |
| Esox lucius | Clauge (Rhône) | BRO427, 428, 430 | A | A | C | G | C | A | C | T | G | A |
| Esox nov. sp. X lucius | Eyre | BRO23 | B | A | C | G | C | R | C | Y | R | A |
| Esox lucius X nov. sp. | Adour | BRO462 | A | A | C | G | C | R | C | Y | R | A |
| Esox lucius X nov. sp. | Lien (Charente) | BRO506 | A | A | C | G | C | R | C | Y | R | A |
| Esox nov. sp. X lucius | Boutonne (Charente) | BRO545 | B | A | C | G | C | R | C | Y | R | A |
| Esox nov. sp. | Estampon (Adour) | BRO531 | B | A | C | G | C | G | C | C | A | A |
| Esox nov. sp. | Geloux (Adour) | BRO534 | B | A | C | G | C | G | C | C | A | A |
| Esox cf. nov. sp. | Lien (Charente) | BRO509 | A | A | C | G | C | G | C | C | A | A |
| Esox cf. nov. sp. | Seugne (Charente) | BRO29 | A | A | C | G | C | G | C | C | A | A |
| Esox nov. sp. | Sonsonnette (Charente) | BRO433 | B | A | C | G | C | G | C | C | A | A |
| Esox cf. nov. sp. | Isle (Dordogne) | FFFtag12251 | A | A | C | G | C | G | C | C | A | A |
| Esox nov. sp. | Eyre | BRO21, 443, 445, 536 | B | A | C | G | C | G | C | C | A | A |
| Esox nov. sp. | Grande Leyre (Eyre) | BRO538, 541 | B | A | C | G | C | G | C | C | A | A |
| Esox reichertii | JN132603 | G | T | A | T | A | T | C | G | G |
Based on the position in the COI tree and the sequence divergence compared to all other pike species, on the distinct nuclear sequences for Plagl2 compared to the other French pikes and E. reichertii, on morpho-meristic characters (see below), and on its distinctness with microsatellite data [15], the divergent pikes are considered as a new pike species Esox nov. sp.
3.3 Molecular confirmation of Esox cisalpinus in France in the 19th century
Twelve samples from historical collections were selected to sequence the short COI fragment. However, only five yielded sequences: two specimens from Lake Mouriscot at Biarritz (Adour drainage, MNHN B-0945), one specimen from Lake Trasimeno in Italy (Tiber drainage, MNHN B-0947), and the two specimens from Lake Geneva at Geneva (Rhône drainage, MNHN B-0949) (Table 2).
This DNA fragment provides a good discrimination of the three species despite its short length (Fig. 2).
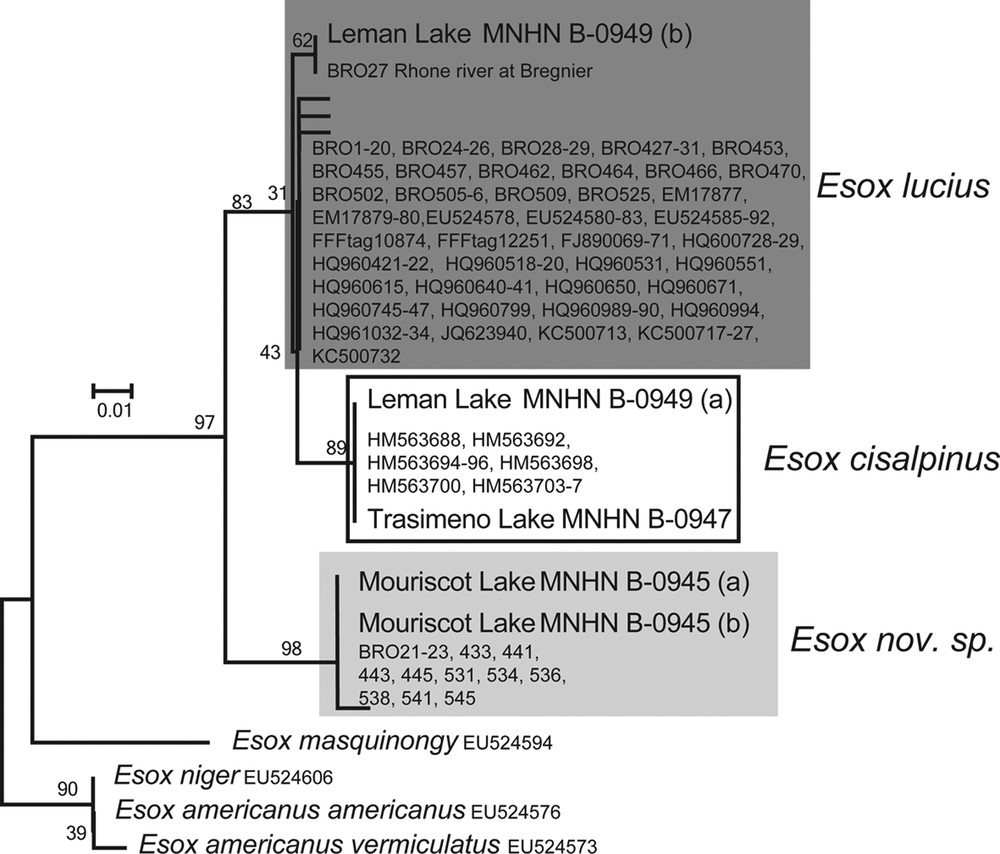
Barcoding NJ K2P tree of 131 bp of the COI marker on 136 individuals of Esox lucius and other esocids, including five specimens from historical collections. The numbers on the nodes represent bootstrap values. A dark grey box highlights Esox lucius samples, a white box E. cisalpinus samples, and a light grey box Esox nov. sp. samples.
In the tree-based identification, all the specimens morphologically assigned to the new species (including the specimens from Lake Mouriscot [MNHN B-0945]) cluster together, and they display six diagnostic sites. These sites are present in the ancient specimens. BLAST-searching the sequences does not yield results with similarity higher than 95% in either BOLD or GenBank. The E. lucius sequences form two clusters because of a single difference. One haplotype is shared between an ancient specimen from Lake Geneva (MNHN B-0949 [b]) and a recent one from the Rhône River, about 80 km downstream (BRO27). All sequences of the specimens identified morphologically as E. cisalpinus cluster together (two diagnostic sites). This cluster includes the specimen from Lake Trasimeno (MNHN B-0947), and the second individual from Lake Geneva (MNHN B-0949 [a]).
Thus, the fragment of COI corroborates the morphological identification of specimens collected more than one century ago. This demonstrates the co-existence of E. lucius and E. cisalpinus in Lake Geneva at this period.
4 Diagnostic description of the Aquitanian pike Esox aquitanicus sp. nov.
4.1 Types and comparative material
The description is based on the observation of six specimens (see below) and completed by the analysis of photographs from six other released individuals. Specimens recognized as hybrids were not included in the description.
Holotype: MNHN 2013-1246, 372 mm SL, female; Adour drainage, Estampon, Saint-Gor; Denys and ONEMA, 08/10/2013. Paratypes: France: MNHN B-0944, 233 mm SL; Charente-Maritime, La Rochelle; D’Enfer, 1824—MNHN B-0945, 2, 207-238 mm SL; Adour drainage, Lake Mouriscot, Biarritz; Blanchard, 1880—MNHN 2013-0838; 231 mm SL; Eyre drainage, Grande Leyre, Sabres; Denys and ONEMA, 10/10/2013—MNHN 2013-1245, 396 mm SL; Eyre drainage, Eyre, Belin-Béliet; Denys, 09/10/2013. Non-type: France: MNHN 2013-0839; 416 mm SL; Charente drainage, Boutonne, Saint-Séverin-sur-Boutonne; Denys, 10/10/2013.
Comparative material. Esox cisalpinus: France: MNHN 2003-0242, 168 mm SL; Rhône drainage, Lake Saint-André; 24/06/1926— Italy: BMNH 1896.10.3.19, 241 mm SL; Po drainage, Lake Garda; Werner—BMNH 1896.10.3.20, 238 mm SL; Po drainage, Lake Garda; Werner— MNHN B-0942, 250 mm SL; Reno drainage, Reno, Bologna; Savigny, 1823—MNHN B-0947, 2, 172–204 mm SL; Tiber drainage, Lake Trasimeno; Canali—MNHN B-0948, 2, 292–341 mm SL; Po drainage, Lake Como; Pentland—Switzerland: MNHN B-0949, 1, 341 mm SL (the second specimen 295 mm SL was not included); Rhône drainage, Lake Geneva, Geneva; Candolle. Esox lucius: BMNH 1853.11.12.114, type possible, 160 mm SL—Canada: MNHN B-2823, 590 mm SL; Saint-Laurent drainage, Lake Erie; Lesueur, 1818—MNHN 1994-0262, 5, 130–243 mm SL; Ontario, Saint-Laurent drainage, Sunshine creek; Momot and Hartviksen, 03/10/1986—Czech Republic: BMNH 1967.8.3.14, 270 mm SL; Danube drainage, Rybnik Vira, Lomnice and Luznici, the pound Vira, about 1 ha, South Bohemia, near the Town Trebon—MNHN 0000-1343, 252 mm SL; Danube drainage, Danube, Olomouc; Jetteleis, 1863—Estonia: BMNH 1925.5.22.17, 196 mm SL; Saadjaro, near Tartu (Dorpat); Piiper—Finland: MNHN 1884-0951, 307 mm SL; Enara; Rabot, 1884—MNHN 1884-0952, 230 mm SL; Enara; Rabot, 1884—MNHN 1884-0953, 335 mm SL; Nota; Rabot, 1884—France: MNHN 0000-1379, 189 mm SL; Seine drainage, Seine; Valenciennes—MNHN A-9974, 321 mm SL; Loire drainage, Lake Grand-Lieu, Grand-Lieu; Thomas, 1858—MNHN B-0946, 206 mm SL; Rhône drainage, Avignon; Blanchard, 1880—MNHN B-0950, 109 mm SL; Rhine drainage, Moselle, Metz; Malherbe, 1850—MNHN 1988-0373, 172 mm SL; Loire drainage, Indre, Le Blizon; Pletikosic and Zimermann, November 1987—MNHN 1993-3498, 285 mm SL; Seine drainage, Seine, Alfortville; Dingerkus and Guilbert, 01/08/1989—MNHN 2003-0133, 185 mm SL; Rhône drainage, Lake Paladru—MNHN 2011-1144, 241 mm SL; Seine drainage, Epte, Guerny; Denys and ONEMA, 20/10/2011—Germany: MNHN B-0943, 274 mm SL; Elbe drainage, Elbe; Nietsch, 1827—Macedonia: BMNH 1928.1.21.1, 263 mm SL; Lake Aghiou Vasseli, near from Salonika—Netherlands: BMNH 1953.6.26.8, 287 mm SL—MNHN 1975-0760, 132 mm SL; Lake Vistonis drainage, Richios; Economidis, 29/05/1975—Russia: MNHN 0000-1706, 2, 343–360 mm SL; Siberia, Irtysh drainage, Irtysh; Humboldt, 1830—MNHN B-0952, 2, 353–366 mm SL; Grande Duchesse Hélène—MNHN 1891-0200, 174 mm SL; Siberia, Petschora drainage, Petschora, Ust Poschow; Rabot, 1891—MNHN 1897-0515, 196 mm SL; Siberia, Ob drainage, Irtych, Kara irtych; Chaffanjon, 1897—MNHN 1897-0516, 155 mm SL; Siberia, Ob drainage, Irtych, Kara irtych; Chaffanjon, 1897—MNHN 1903-0141, 272 mm SL; Lake Baikal; Labbe, 1903—Switzerland: MNHN B-0941, 347 mm SL; Rhine drainage, Lake Zug; Major—United-States: MNHN A-0793, 850 mm SL; Mississippi drainage, Wabash; Lesueur—MNHN A-1272, 151 mm SL; Lac Michigan, Oconomowoe; Jordan, 1879.
4.2 Diagnosis
Esox aquitanicus is distinguished from the two other European species by a color pattern of the sides, with 1–1.5 scale wide oblique vertical bars conferring it a marbled coat (Fig. 3) and the combination of the following morpho-meristical characters: a shorter head with a snout 0.9 times larger than postorbital length (Fig. 4), an anal fin basis 1.1–1.2 times larger than caudal peduncle length, mainly 101 to 121 lateral scales and 53 to 57 vertebrae.

(Color online.) Holotype of Esox aquitanicus MNHN 2013-1246, 372 mm SL, coloration alive (a), and after fixation in formalin (b).

(Color online.) Head profile showing the difference in snout size of two juveniles caught in July of their birth year at one day interval: Esox aquitanicus BRO443, Eyre at Bélin-Béliet (a), and Esox lucius BRO453, Isle (Dordogne drainage) at Saint-Médard-de-Guizière (b); the specimens were released alive; scale bar: 30 mm.
4.3 Description
The general appearance is shown in Figs. 3 and 4a; morphometric data are given in Table 4. The holotype counts are given first, followed by the paratype counts in brackets, if different.
Morphometry of Esox aquitanicus, E. cisalpinus and E. lucius. Values in parentheses: mean. Values of holotype included in range. Bold mean values highlight significant morphometric differences.
| E. aquitanicus | E. cisalpinus | E. lucius | ||
| Number of specimens | 6 | Holotype | 7 | 38 |
| Standard length (mm) | 207–396 | 372 | 168–341 | 106–850 |
| In percent of standard length | ||||
| Head length | 28.6–34.3 (32.1) | 28.6 | 31.8–33.8 (33.0) | 27.3–37.1 (32.4) |
| Predorsal length | 72.6–79.8 (77.0) | 72.6 | 75.7–79.0 (77.8) | 73.7–79.7 (76.5) |
| Prepectoral length | 24.7–30.0 (27.8) | 24.7 | 27.7–31.1 (28.8) | 23.2–32.4 (28.2) |
| Prepelvic length | 51.5–59.9 (55.8) | 51.5 | 54.4–59.6 (56.9) | 52.6–60.7 (56.4) |
| Preanal length | 74.7–83.6 (79.4) | 74.7 | 77.5–83.1 (80.6) | 76.3–84.1 (79.5) |
| Post-dorsal length | 8.0–12.2 (10.3) | 12.1 | 8.5–11.2 (9.8) | 8.4–12.5 (10.5) |
| Length of caudal peduncle | 9.0–10.9 (10.1) | 10.8 | 8.5–11.7 (10.1) | 7.9–13.7 (9.9) |
| Distance between pectoral and pelvic fin bases | 24.2–31.6 (26.9) | 24.5 | 24.6–29.3 (27.0) | 22.7–31.9 (26.8) |
| Distance between pelvic fin base and anal fin origin | 20.3–24.0 (22.0) | 20.9 | 19.5–23.8 (21.7) | 18.1–25.4 (21.6) |
| Length of dorsal fin basis | 13.3–15.5 (14.1) | 14.3 | 12.2–14.3 (13.1) | 11.5–15.7 (13.7) |
| Length of pectoral fin | 11.3–14.9 (13.7) | 11.3 | 11.4–14.7 (13.1) | 8.8–15.9 (12.8) |
| Length of pelvic fin | 11.0–14.5 (13.4) | 11.0 | 11.8–14.2 (13.0) | 8.6–14.9 (13.1) |
| Length of anal fin basis | 11.2–12.1 (11.7) | 11.7 | 9.2–11.4 (10.3) | 7.7–12.0 (10.4) |
| Body depth | 16.9–18.6 (17.8) | 17.3 | 15.2–20.3 (17.4) | 13.8–19.7 (16.4) |
| Depth of caudal peduncle | 7.2–8.3 (7.6) | 7.2 | 6.3–7.9 (7.0) | 5.1–8.1 (6.7) |
| In percent of head length | ||||
| Snout length | 39.2–42.3 (40.7) | 42.3 | 40.7–47.8 (43.1) | 39.8–52.1 (43.5) |
| Postorbital length | 42.3–46.3 (44.1) | 45.1 | 38.4–43.7 (41.8) | 36.7–46.7 (41.4) |
| Eye diameter | 11.8–17.5 (15.2) | 12.6 | 11.7–17.2 (15.1) | 8.6–22.3 (15.1) |
| Interorbital width | 17.2–22.9 (20.5) | 22.5 | 14.9–20.0 (17.8) | 13.8–23.7 (19.1) |
This species has an extended and spindle-shaped body, a single dorsal fin in backwards position and opposed to the anal fin. Its large and flattened snout is 42.3% (39.2–42.3%) larger than head length. Its mandible is longer than the upper jaw, and its teeth are implanted on vomer, tongue, palatins and inter-maxillaries. Small cycloid scales are present on preopercula and on the upper half of the opercula. The anal fin basis is 11.7% (11.2–12.1%) larger than standard length. The depth of caudal peduncle is 7.2 (7.3–8.3%) larger than standard length.
Lateral line with 106 (103–108) (101–121 with additional data from released samples) scales (Fig. 5); 12 (10–13) scales rows between the basis of pelvic fins and lateral line; 11 (11–13) scales rows between the basis of dorsal fin and lateral line. Dorsal fin with 6 (6–7) simple rays and 15 (13–15) branched rays. Anal fin with 4 (3–4) simple rays and 12 (12–14) branched rays. Pectoral fins with 1 simple ray and 14 (12–15) branched rays. Pelvic fins with 1 simple rays and 10 (9–10) branched rays. Caudal fin forked with 19 rays. Branchiostegal rays: 14 (13–14). Submandibular pores: 5–5. Vertebrae: 55 (53–57).
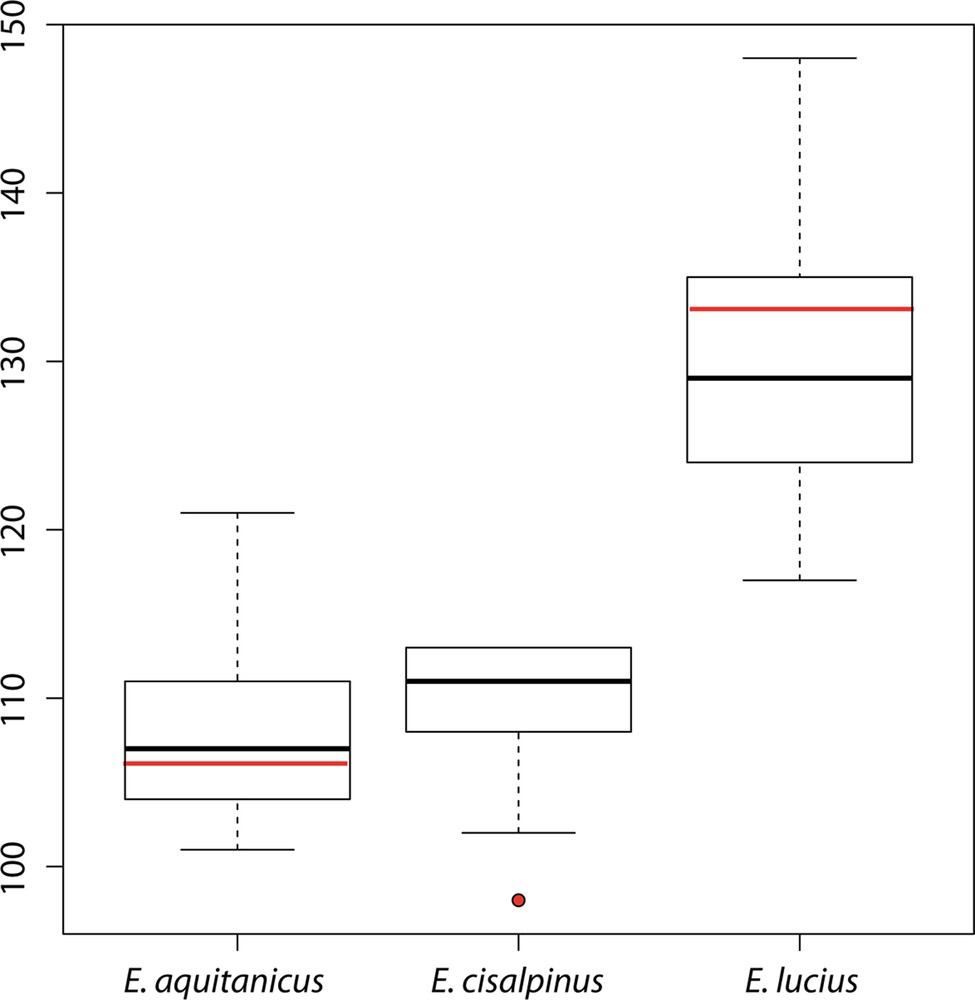
(Color online.) Boxplot representing the total number of scales on lateral line of Esox aquitanicus (n = 13), E. cisalpinus (n = 9), and E. lucius (n = 50). Red marks represent type specimens.
4.4 Comparison with closely related species
Esox aquitanicus is distinguished from E. lucius (from any origin) by 24 diagnostic sites on the long mitochondrial COI dataset and 3 in the nuclear Plagl2 dataset, by having fewer lateral scales (101–121, vs. 117–148; Fig. 5), a shorter snout (average 40.7% HL, vs. 43.5% HL; Fig. 4), a longer postorbital length (average 44.1% HL, vs. 41.4% HL), a slightly longer basis of the anal fin (average 11.7% SL, vs. 10.4% SL), a slightly deeper caudal peduncle (average 7.6% SL, vs. mean 6.7% SL), and a lower number of vertebrae (53–57, vs. 57–65; [20]).
Esox aquitanicus is distinguished from E. cisalpinus by 26 diagnostic sites on the long COI dataset, by having a shorter snout (average 40.7% HL, vs. 43.1% HL), a longer postorbital length (average 44.1% HL, vs. 41.8% HL), a larger interorbital width length (average 20.4% HL, vs. 17.8% HL), a longer basis of the anal fin (average 11.6% SL, vs. 10.3% SL), a slightly deeper caudal peduncle (average 7.6% SL, vs. 7.0% SL), and a slightly higher number of branchiostegal rays (13–14, vs. 12–13).
Thus, Esox aquitanicus differs from both species by having a snout shorter than the preopercular length (ratio snout length/preopercular length: 0.9%, vs. 0.9–1.3%), an anal fin basis always larger than the caudal peduncle (ratio anal fin basis/caudal peduncle length: 1.1–1.2%, vs. 0.7–1.4%), and by a marbled coat with 1–1.5 scale wide oblique vertical bands tending, in larger specimens, to be discontinuous until they form little irregular white blotches. The comparison was also made with specimens measuring between 200 and 400 mm SL (representing 60% of our sampling), and the conclusion was the same. As shown in Fig. 4 with two juveniles caught in July of their birth year at one day interval, the difference of snout size is not due to an allometric effect.
4.5 Color in life
Esox aquitanicus has grey to yellow–green flanks adorned with 16 to 30 oblique vertical bars with a width of 1–1.5 scale (very well marked in juveniles), which tend to be discontinuous into little irregular white blotches in larger fish, conferring a marbled aspect with very small white blotches (Fig. 3a). The fins’ color is yellow to orange. Dark pigmentation on paired fins are faint, as opposed to the unpaired fins which have well-developed dark vermiculations. Like E. lucius and E. cisalpinus, young individuals have a contrasting brownish vertical bar under the eye.
4.6 Color in preservation
Samples preserved in formalin have a brown coloration, and the light parts like the belly and the ornamentation are yellow. Fins lose their pigmentation but not their dark blotches; only one specimen has conserved a trace of its orange pigmentation in the caudal fin after one month (MNHN 2013-0878). The brownish bar under the eye is still visible (Fig. 3b).
4.7 Ecology and notes on biology
E. aquitanicus should have similar ecological characteristics as E. lucius in terms of habitat, behavior, and predation [41], because no difference were mentioned about ecological traits between Aquitanian and the other French pike populations, except an earlier spawning in February instead of March–April [23]. Its size can exceed 1000 mm TL (Dégrave, pers. comm.) A pike of 1370 mm was mentioned by Laporte [42] in Lake Cazeau, right in the heart of the distribution area of E. aquitanicus. However, we cannot exclude early stockings of E. lucius. When there is cohabitation between the two species, the Aquitanian pike is able to hybridize with the northern pike (present study).
4.8 Distribution
The Aquitanian pike is present in the Charente, Dordogne, Eyre, and Adour basins. Lake Mouriscot constitutes its currently known most southern location (Fig. 6; [23]). Considering the biogeographical history of Atlantic French drainages [43], it should occur in the Garonne basin too, but the population could have regressed following the introduction of the Northern Pike E. lucius. There are still higher probabilities to encounter the Aquitanian pike in small tributaries, where humans do not stock Northern Pike, than in larger streams. It might be present in the “Sèvre niortaise” basin, but no specimen was captured, and in the Loire drainage, but we captured only E. lucius individuals. It is possible that Aquitanian pike is stocked and sold as E. lucius by fish farmers of southwestern France, in order to restock waterbodies in France or abroad for sport fishing. Additional investigations to characterize the limits of the distribution area are necessary.
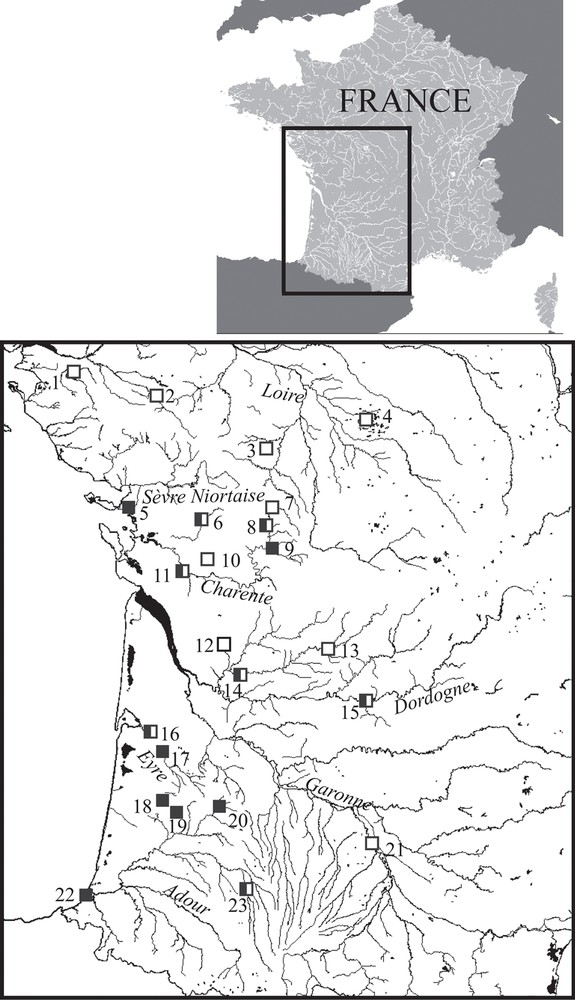
Occurrences of Esox aquitanicus (black squares) and E. lucius (white squares) in the Southwest of France; numbers indicate the following locations: Lake Grand-Lieu (1), Sèvre nantaise at St-Malo-du-Bois (2), Boivre at Béruges (3), Indre at Le Blizon (4), La Rochelle (5), Boutonne at Saint-Séverin-sur-Boutonne (6), Charente at Saint-Saviol (7), Lien at Condac (8), Sonsonnette at Saint Front (9), Antenne at Le Seure (10), Seugne at Les Gonds (11), Lary at St-Martin-d’Ary (12), Isle at Trélissac (13), Isle at St-Médard-de-Guizière (14), Dordogne at Cénac-et-Saint-Julien (15), Eyre at Mios (16), Eyre at Bélin-Béliet (17), Grande Leyre at Sabres (18), Geloux at Garein (19), Estampon at Saint-Gor (20), Garonne at Verdun-sur-Garonne (21), Lake Mouriscot at Biarritz (22), Adour at Estirac (23).
4.9 Etymology
The specific name aquitanicus is the adjective of Aquitania referring to the region of southwestern France, Aquitaine, where the species was discovered. For this reason, the vernacular name chosen is “Aquitanian pike” (“brochet aquitain” in French).
4.10 Molecular reference
Table 5 lists the GenBank accession numbers of COI and Plagl2 DNA sequences from specimens for which morphological and molecular data are available.
DNA sequences of COI and Plagl2 and vouchers characterizing Esox aquitanicus. The sequences were tagged as suggested by the nomenclature of Chakrabarty et al. [46].
| Specimen Catalog | Tissu Catalog | (provisional) GenBank Accession Number | GenSeq Nomenclature | |
| COI | Plagl2 | |||
| MNHN 2013-1246 | MNHN ICTI 6388 | BRO531 | BRO531 | Genseq-1 COI, Plagl2 |
| MNHN 2013-1245 | MNHN ICTI 6389 | BRO536 | BRO536 | Genseq-2 COI, Plagl2 |
| MNHN 2013-0878 | MNHN ICTI 6390 | BRO538 | BRO538 | Genseq-2 COI, Plagl2 |
| MNHN B-0945 | MNHN ICTI 6391 | MNHN B-945a | NA | Genseq-2 COI |
| MNHN B-0945 | MNHN ICTI 6392 | MNHN B-945b | NA | Genseq-2 COI |
| photo voucher | MNHN ICTI 6393 | BRO443 | BRO443 | Genseq-5 COI, Plagl2 |
| photo voucher | MNHN ICTI 6394 | BRO445 | BRO445 | Genseq-5 COI, Plagl2 |
| photo voucher | MNHN ICTI 6395 | BRO534 | BRO534 | Genseq-5 COI, Plagl2 |
| photo voucher | MNHN ICTI 6396 | BRO541 | BRO541 | Genseq-5 COI, Plagl2 |
5 Discussion
5.1 Evidence for three species of pike in France
Morpho-meristical analysis discriminates Esox lucius and a group of pikes from southwestern France (see below). Moreover, the partial sequence for the mitochondrial gene COI for these specimens diverges from sequences for E. lucius by 4.0%. Most divergences between North American pike species are above 2% (except between E. niger and E. americanus americanus) [44,45]. The nuclear marker Plagl2 corroborates this result. Our genetic analysis is in agreement with the microsatellite study of Launey et al. [15] that included specimens from France and other European countries. These independent datasets all point to the existence of a new species of pike Esox aquitanicus, from Charente to Adour drainages.
Descriptions of E. cisalpinus/E. flaviae both lack complete morphometric studies. There is also a difference between the lateral scales numbers in the two descriptions: 101–115 [6] vs. 92–107 [18]. In our study, specimens of this species have 102 to 113 lateral scales. E. cisalpinus samples form a cluster included within the E. lucius clade in the COI dataset analyzed by BI, whereas the NJ distance analysis separates them in distinct clusters. There are diagnostic sites for this cluster in the COI sequence, so the assignation of specimens to E. cisalpinus or E. lucius is possible. Other molecular markers also support the distinction between these two species [6,13,14], justifying thus the recognition of E. cisalpinus as a distinct evolutionary unit. Because all our specimens came from old collections, no Plagl2 sequence could be obtained for E. cisalpinus in this study. It would be interesting to get some nuclear sequences of this species.
Two groups of COI haplotypes are present in French E. lucius: one closest to Elbe and Danube drainages, and the other closest to Canadian populations. French specimens associated with the “Canadian group” might have an aquaculture origin (probably North American). Miller and Senanan [12] observed genetic differentiation between North American and European pike populations, and recommended avoiding transcontinental restocking in order to maintain the genetic characterization. Our results indicate that such introductions probably took place in France and in other European drainages from North America or vice-versa. However, another freshwater fish species, actually better salt tolerant, the three-spined stickleback Gasterosteus aculeatus Linnaeus, 1758 also displays such a similarity between both sides of the Atlantic Ocean [47].
The molecular identification checked with the morpho-meristic characters of old collections samples held at the MNHN uncovered specimens belonging to the three species. Out of two specimens caught in Lake Geneva in the first half of the 19th century (MNHN B-0949), one was identified genetically as E. cisalpinus, whereas the other one was determined as E. lucius. Lake Geneva borders Switzerland and France, so this analysis is the first known occurrence of E. cisalpinus in France, one century before the first recorded pike restocking campaign [48]. It confirms the presence of this species in the peri-alpine lakes north of the Alp range axis. These two species co-occurred during this period. Nicod et al. [13] sampled for their study seven specimens of pike in Lake Geneva at Geneva, 4 in Lake Bourget, which is close to Lake Saint-André. All samples present the E. lucius haplotype rather than the E. cisalpinus haplotype. It would however be necessary to check again for E. cisalpinus in all the peri-alpine lakes on a larger number of samples, investigating more precisely possible traces left in the genomes of specimens appearing as Esox lucius. Pike populations were very abundant in the peri-alpine lakes during the 16th century [49], but pike restocking became quite frequent in Switzerland during the 20th century [50], and this has had an impact on the genetic variability of native populations [15].
5.2 Hybridization with the northern pike
Hybridization is common within freshwater fishes in many groups, including Esocid species [51]. While the hybrid E. lucius × masquinongy is sterile, the hybrid of the two more closely related E. lucius × reichertii is fertile [1]. E. lucius is more closely related to E. aquitanicus (and even more to E. cisalpinus) than to E. reichertii. An old occurrence of introgressive hybridization is also suspected within Esocids between E. niger and E. americanus americanus [45].
Lucentini et al. [6] pointed out cohabitation of E. lucius and E. cisalpinus in northern Italy, and hypothesized that specimens combining lower lateral scale numbers with the color pattern of northern pikes are their hybrids. One of the MNHN pikes from Lake Geneva was identified genetically as E. Lucius, but has a low lateral scales number (113); this might result from hybridization between cohabiting E. cisalpinus and E. lucius.
Plagl2 sequences discriminate E. aquitanicus from E. Lucius, with three diagnostic sites. The Plagl2 gene is a nuclear marker, and some specimens from Adour, Charente and Eyre drainages are heterozygous, with both alleles present. Moreover, some specimens from these same basins were identified morphologically and with COI as one species, though they are heterozygous, with the haplotypes of both species present. It is the case of our two specimens caught in the Charente drainage (MNHN 2013-1247) which have the Plagl2 allele of E. aquitanicus (BRO506 and BRO509 being respectively heterozygous and homozygous), and an association of morphological characters from to the two species; they could therefore be considered as potential hybrids. Nevertheless more genetic studies with additional nuclear markers are needed in order to confirm this hypothesis [52].
5.3 Biogeography and conservation
The Northern Pike populations in France are regressing because of pollution, over-fishing, bank harnessing and dams, and the lost access to spawning grounds. For these reasons, this species is classified as “vulnerable” on the French red list of threatened species [20,53].
The Aquitanian pike E. aquitanicus endemic from Charente to Adour drainages, like several other species in the Southwest of France [16,20] has a much smaller distribution area. Moreover, further introductions of Northern Pike from fish farming in eastern France and other areas [23,54], induce both hybridization and competition, and hybridization resulting from introduction of a non-native species is a major threat to endemic freshwater fishes [55]. Studies are strongly required to better assess the actual status and distribution of this new species.
The Cisalpine Pike E. cisalpinus is native to Italian drainages. Nevertheless, to our knowledge the Rhône drainage was never connected with any Adriatic basin. Lake Geneva was somewhat connected with the Rhine drainage during the retreat of the huge Rhône glacier that covered most of the Swiss plateau at the end of the last glacial maximum, and at the former glacial maximums probably as well [43]. Only cold water fish fauna like salmonids were able to take advantage of this opportunity to colonize Lake Geneva. While pikes do not fear cold water, we do not know whether it was able to swim back upstream to reach Lake Geneva, as this depended on the unknown water velocities prevailing at the northern outlet of the lake. Similarly, despite its presence in the French Rhône drainage basin during Paleolithic and Neolithic ages [56], there is no evidence that it had the opportunity to cross the so-called “Pertes du Rhône” waterfall upwards between Lake Geneva upstream and Lake Bourget downstream. Thus, the Cisalpine Pike is probably not native to Lake Geneva, and could be the result of an ancient introduction: pike is already mentioned as a duty of the Geneva monastery to the Aosta church in Italy around 1150 [57]. In Italy, the status of E. cisalpinus is still ‘data deficient’, but Bianco [19] considers it ‘vulnerable’ because of the possibility of hybridization with E. lucius.
Because pike is a very important recreational and commercial fish, in France and in other countries, it is highly manipulated and populations are regulated by fish dumping. Where only one species was previously known in France, our study highlights the presence of two other species with little known distributions. Further investigations are necessary to explore their biogeography, and their ecological traits. Both appear to occupy limited areas, and are threatened by the introduction of the Northern Pike E. lucius. They therefore deserve a place in the red list of threatened freshwater fish in France [58,59], in order to organize the first conservation measures.
Acknowledgements
This work was supported by the “Muséum national d’histoire naturelle”, the UMR BOREA and the French “Office de l’eau et des milieux aquatiques” (ONEMA). We are particularly grateful to N. Poulet and S. Dembski. We thank the “Fédération de la pêche” of Charente and all the ONEMA agents (especially M. Goillon, R. Martin, S. Vincelot, S. Manné, and S. Mougenez), as well as G. Hautecœur and the Pêcherie Parpillon for fish samplings. We also thank L. Dégrave (“Parc naturel régional des Landes”), and S. Lefebvre (ONEMA) for their useful information. The ichthyology curators of MNHN and BMNH gave access to the specimens, and provided the X-ray pictures. M. Mennesson, R. Debruyne and C. Bonillo helped with laboratory work. Laboratory access and assistance was provided by the “Service de systématique moléculaire” of the “Muséum national d’histoire naturelle” (CNRS UMS 2700). And finally, we are grateful to J. Allardi, G. Carrel, A. Hassanin, and F.J. Meunier for their advice and G. Lancelot and N. Schnell for their help with the manuscript.


