1 Introduction
Rice is the most important food crop for over half of the world's population [1] and supplies 20% of daily calories [2]. Among the biotic stresses, blast disease is the most-destructive disease in rice [3]. Still now, developing blast-resistant varieties is the most cost-effective method to improve rice blast disease resistance in rice [4]. MR219 is a Malaysian rice variety having high-yield potential (10.75 tons ha−1) with a short maturation period (105 to 111 days) and good eating quality [5]. Nowadays, MR219 is grown in more than 90% of rice fields in Malaysia due to its high production [6]. When MR219 was released by the Malaysian Agricultural Research and Development Institute (MARDI) in 2001, it was moderately resistant against blast fungal Magnaporthe oryzae. However, after a long time cultivation and regeneration of seeds and due to environmental changes, new blast pathotypes have been developed, which is why the blast-resistant ability of MR219 is adrift and affected severely with blast pathogens. The main objective of this research was to convert the widely cultivated variety MR219 into a blast-resistant variety with MR219 genetic backgrounds through marker-assisted backcrossing and marker-assisted selection (MAS) using Malaysian local rice variety Pongsu Seribu 1, which is resistant against blast pathogen M. oryzae. The utilization of blast-resistant genes in breeding is the most effective and economic strategy for controlling rice blast.
Recent advancements in biotechnology have led to the development of more efficient selection tools to substitute phenotype-based selection systems for varietal development [7]. Marker-assisted selection has been successfully utilized for developing improved parental lines of hybrid rice with inbuilt resistance to bacterial blight and blast diseases [8,9]. MAS is a kind of breeding selection for target genes through analysis with molecular markers closely linked to the target gene. The marker-assisted selection results are considered as higher reliability because of their selective effects, which are independent of gene effects and environmental factors. Using MAS, genotype selection could be performed in early generation, thus speeding up the breeding program [10]. The selection efficiency increased greatly with the utilization of DNA markers in our marker-assisted backcrossing (MABC) program. The MABC approach develops an ideal genotype within a short time avoiding the complicated issues related to transgenic technology and conventional breeding approaches [11]. The tremendous benefits of MABC are:
- • competent foreground selection for the locus of interest;
- • effective background selection for the recovery of recurrent parent genome;
- • reduce linkage drag adjacent to the introgressed locus;
- • prompt breeding for the development of new genotypes with favorable traits.
The efficiency of MABC depends upon the availability of tightly linked markers or flanking markers for the target locus, population size, the position and number of background markers and the number of backcrosses [12]. MABC has previously been used in rice breeding to incorporate the bacterial blight resistance gene Xa21 into elite cultivars [13]. Employing molecular markers with known map position speeds up recovery of the recurrent parent genome (RPG) by about two to three generations [14,15]. Although the extent of effectiveness of this program is delimited by some factors, molecular breeding technologies, which have already been proven as the most effective ones for the development of any resistant variety [16], are upgrading day by day. The general objective of the backcross breeding method is to retain the gene(s) of interest and to eliminate the remainder of the donor parent genome as rapidly and efficiently as possible. With each successive backcross, the average percentage of the genome of the offspring derived from the donor line decreases and the percentage of the total recurrent parent genome increases [17].
Marker-assisted foreground selection and stringent phenotypic selection in each crossed generation ensure selection of plant with desirable alleles, which has maximum recurrent parent phenome recovery [18]. As a general rule, two to four markers per 100 cM can be efficiently used to accelerate the recovery of the RP in the early generations such as BC1 or BC2 [14,19]. The objective of the background selection is to accelerate the return to the recipient parent genome outside the target gene, in addition to knowing how much recurrent parent genome is recovered in the subsequent generation using marker-assisted backcross breeding program. The objective of this study was to calculate the recurrent parent genome recovery contribution in marker-assisted backcross breeding programs for the introgression of blast-resistant gene into a rice cultivar derived from MR219 and Pongsu Seribu 1-rice variety.
2 Materials and methods
2.1 Plant materials and breeding strategy
MR219 was used as the recurrent parent for incorporation of blast resistance gene(s) within the donor Pongsu Seribu 1, which is a Malaysian local rice variety resistant to blast. The recurrent parent MR219 was crossed as the female parent with the donors, Pongsu Seribu 1. The two-F1 plants positive for Pi-gene(s) were backcrossed with MR219 to produce the BC1F1 seeds. Foreground selection in the BC1F1 generation for the blast-resistant genes was performed using tightly linked markers (RM6836 and RM8225). The gene positive plants with the highest background recovery and maximum phenotypic similarity to the recurrent parent were backcrossed with MR219 to generate BC2F1 seeds in each backcross. Foreground, background and phenotypic selections were carried out to select elite plants from each backcross series.
2.2 Molecular marker analysis
The rice SSR markers, which were found as tightly linked to Pi-gene, were used for foreground selection. Marker polymorphism survey was carried out between the two parents using foreground markers (RM168, RM413, RM5961, RM8225, RM6836, RM224, RM140, RM101, RM247, RM261, RM340, RM547, RM495, RM251, RM229 and RM5), which have been found [20] to be related to Pi-genes. The positions of these markers are given in Table 1.
Information of polymorphic markers used in this study.
| Polymorphic SSR markers | Chromosome position |
| RM5, RM140, RM495, RM3252, RM4959, RM10022 | 1 |
| RM262, RM573, RM1234, RM1358, RM5529 | 2 |
| RM7, RM36, RM168, RM218, RM251, RM3131 | 3 |
| RM252, RM261, RM280, RM348, RM471, RM5633, RM8213 | 4 |
| RM413, RM480, RM1054, RM1089, RM1187, RM1237 | 5 |
| RM193, RM340, RM508, RM528, RM586, RM587, RM3827, RM6836, RM8225, RM19311 | 6 |
| RM11, RM336, RM1132, RM1357, RM1362, RM1364 | 7 |
| RM85, RM152, RM342, RM547, RM1109, RM1111, RM1235 | 8 |
| RM205, RM3331, RM3609, RM23697 | 9 |
| RM285, RM1375, RM2863, RM3123 | 10 |
| RM224, RM229, RM286, RM5961 | 11 |
| RM101, RM247, RM3813, RM5479, RM7018 | 12 |
A set of 375 SSR markers uniformly spread across the 12-rice chromosomes were screened for polymorphism survey between the recurrent parent, MR219, and the resistant donor, Pongsu Seribu 1, to find out polymorphic background markers. The primer sequences for SSR markers were adapted from the Gramene SSR marker resource (http://www.gramene.org/). At least four polymorphic SSR background markers per chromosome were used for background selection to recover the recipient genome.
2.3 DNA isolation, PCR conditions and electrophoresis
The total genomic DNA was extracted from fresh leaves of 4-week-old individual plants using the CTAB method [21] with some modifications. DNA was quantified by using nano-drop spectrophotometry (ND1000 Spectrophotometer). The diluted DNA samples were again diluted with 1× TE buffer (10 mM Tris-HCl, pH 8.0, 1 mM EDTA, pH 8.0) to get a concentration of 70 ng/μL and kept in the refrigerator of −20 °C for PCR analysis.
The total PCR reaction volume of 15 μL contained 70 ng of template DNA, 1.0 μM of each primer, 7.4 μL of master mix (Thermo Scientific) and 4.6 μL of nuclease-free water. In the case of foreground markers, PCR amplification was carried out in a thermocycler (T100™, Bio-Rad), using an initial denaturation at 94 °C for 5 min followed by 35 cycles at 94 °C for 30 s, 55 °C for 30 s, 72 °C for 30 s, the final extension at 72 °C for 5 min, followed by rapid cooling to 4 °C prior to analysis. For background marker, PCR amplification was carried out using touch-down PCR program with the following profile: 94 °C for 3 min, followed by 10 cycles of 94 °C for 30 s, 62 °C for 1 min (decreasing 1 °C per cycle), and 72 °C for 30 s, and 30 cycles of 94 °C for 30 s, 52 °C for 1 min, 72 °C for 2 min, and a final extension at 72 °C for 10 min by rapid cooling to 4 °C prior to analysis. For electrophoresis, a 3.0% metaphor™ agarose (Lonza) gel was prepared, containing 1 μL of Midori green in a 1× TBE buffer (0.05 M Tris, 0.05 M boric acid, 1 mM EDTA, pH 8.0). The gel was run at a constant voltage of 80 V for 80 min and the resolved amplified products were visualized using Molecular Imager® (GelDoc™ XR, Bio-Rad Laboratories Inc., USA).
2.4 Phenotypic selection
The plants having blast-resistant gene(s) with maximum phenotypic resemblance to the recipient parent MR219 were selected at vegetative and flowering stages. Phenotypic selection was carried out over the entire population of BC1F1, BC2F1 and BC2F2 generations after foreground selection. The phenotypic parameters considered at the vegetative stage included plant height, tillering patterns, number of tillers per hill, leaf size, leaf shape and leaf angle, as suggested in [22]. The additional parameters considered at the flowering stage included the panicle shape, panicle angle to the axis, spikelet shape, spikelet size, flag leaf size, and shape. The selected plants were ranked based on their degree of phenotypic resemblance with MR219, and backcross seeds were produced from the individuals with the highest phenotypic rankings. In every generation, the same approach was followed for the selection of individual plants with blast-resistant gene.
2.5 Statistical analysis
The primers banding pattern was scored with reference to two parents. In the case of foreground selection, the band having same level of Pongsu Seribu 1 was scored as ‘R’, which indicated the homozygous allele of the resistant parent for a particular SSR marker. Similarly, the bands with the similar level of MR219 was scored as ‘S’. The Chi2 analysis for the susceptible and resistant ratio was calculated by using the formula, Chi2 = (O − E)2/E, where O is the observed value, and E is the expected value. In the case of background selection, the marker data was analyzed using Graphical Genotyper (GGT 2.0) software [23]. The homozygous recipient allele, homozygous donor allele, and heterozygous allele were scored as “A”, “B”, and “H”, respectively. The Microsoft Excel® file containing these data was imported into an Excel data exchange of GGT 2.0-software program for further analysis. The percentage of markers homozygous for recipient parent (%A), the percent donor alleles (%B) and heterozygous plants (%H) were calculated. The mean difference between morphological data from the parental lines and blast-resistant improved lines were analyzed by independent t-test using SPSS 16.0 program.
3 Results
3.1 Markers polymorphism in the parental line
All tightly linked SSR markers showed clear polymorphisms in the parental line. For background study, out of 375 SSR markers, 70 SSR markers covering all 12-rice chromosomes were identified as polymorphic markers between the two parents (Table 1). The ratio of polymorphic markers on parental survey is approximately 18.67%. All these markers were used in the BC1F1, BC2F1 and BC2F2 generations for background analysis.
3.2 Genotyping F1 generation
The F1 seeds were produced from the cross MR219 and Pongsu Seribu 1, and F1 plants were tested for hybridity using the tightly linked polymorphic foreground SSR markers. In F1 generation, all plants showed heterozygous condition using the foreground markers.
3.3 Genotyping BC1F1 generation
3.3.1 Marker-assisted foreground selection
The best two gene positive F1 plants were backcrossed with MR219 to generate 100 BC1F1 plants. Of the 100 BC1F1 plants, 58 plants were heterozygous for the markers RM6836 and RM8225 tightly linked to Pi-genes (Fig. 1). Plants with heterozygous condition were not identified by using the other foreground markers, indicating that some blast-resistant genes disappeared due to the backcrossing with MR219. The proportion of susceptible and resistant plants in BC1F1 generation using foreground marker are shown in Table 3. The Chi2 results fitted to the expected 1:1 ratio of BC1F1 generation (Chi2 = 2.26), which is non-significant at a probability level of 0.05 (Table 3).
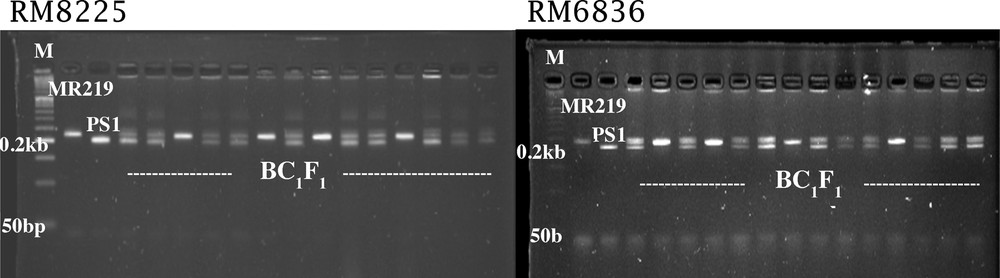
Resistant and susceptible plant screening using RM8225 and RM6836 marker in BC1F1 population.
Proportion of susceptible and resistant genotypes in BC1F1 and BC2F1 generations.
| Generation | Number of line | Observed line | Expected ratio | Chi2 value | P value | |
| Resistant | Susceptible | |||||
| BC1F1 | 100 | 58 | 42 | 1:1 | 2.26 | 0.1328 |
| BC2F1 | 333 | 159 | 174 | 1:1 | 0.58 | 0.4463 |
3.3.2 Marker-assisted background selection
The number of polymorphic markers per chromosome ranged from 4 (chromosomes 9, 10 and 11) to 10 on blast-resistant gene carrier chromosome 6. A total of 70 SSR markers were used for background analysis in 58-BC1F1 plants resulting from foreground selection. The recurrent parent genome (RPG) recovery ranged from 75.40% to 91.3% in BC1F1 generation (Fig. 2).
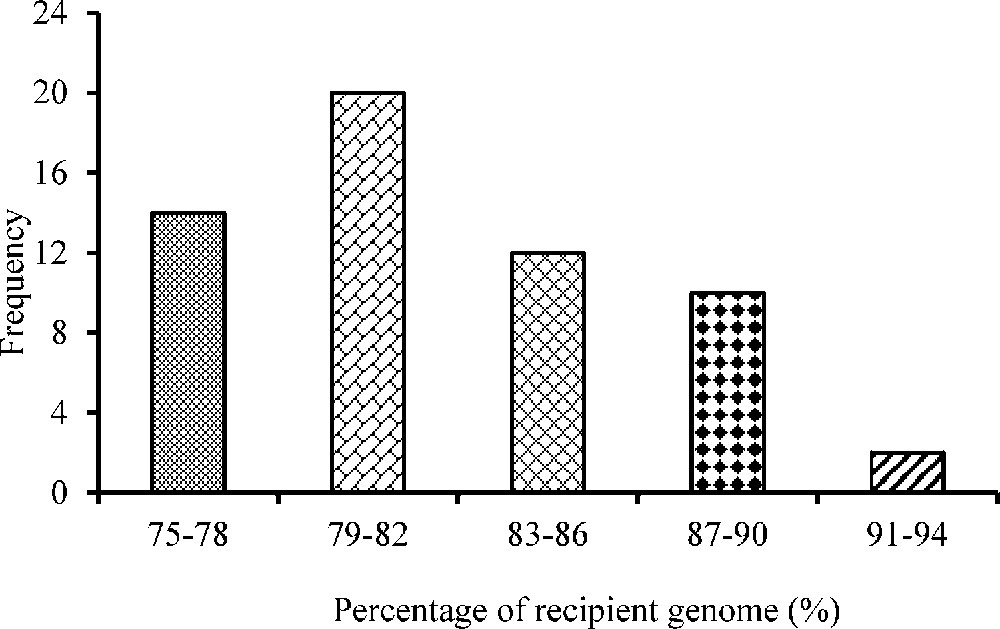
Frequency distribution of the recurrent parent genome recovery (%) in BC1F1 population.
Average RPG recovery of selected 4 BC1F1 plants was 87.3%. The summary result of RPG recovery and heterozygous segment of selected individuals in BC1F1 population is presented in Table 2. The blast-resistant gene donor parent segments appeared on chromosome 6 (Fig. 3). It was observed that chromosomes 1, 8, and 11 were fully recovered in all of the selected lines as compared to other chromosomes (Fig. 3). The best individual in BC1F1 generation was plant No. 6-4, having the highest recurrent parent genome recovery (91.3%) and the lowest heterozygous segments. Chromosome-wise recurrent parent genome recovery of this plant was shown in Fig. 4. Based on the foreground and background selection, four selected BC1F1 plants were used to develop BC2F1 populations.
Background and introgressed segment analysis in selected lines of BC1F1 population.
| Selected individual | A (%) | B (%) | H (%) | Total (cM) | H-segments |
| 6-1 | 86.9 | 1.6 | 11.5 | 1162.3 | 11 |
| 6-2 | 87.2 | 1.6 | 11.2 | 1162.3 | 11 |
| 6-3 | 83.9 | 1.6 | 14.5 | 1162.3 | 8 |
| 6-4 | 91.3 | 1.6 | 7.1 | 1162.3 | 8 |
| Average | 87.3 | 1.6 | 11.08 | 1162.3 | 9.5 |
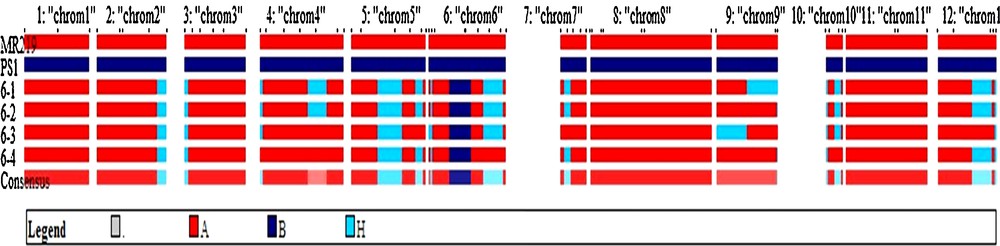
(Color online.) Chromosome-wise recurrent genome recovery of the four selected plants in BC1F1 generation.
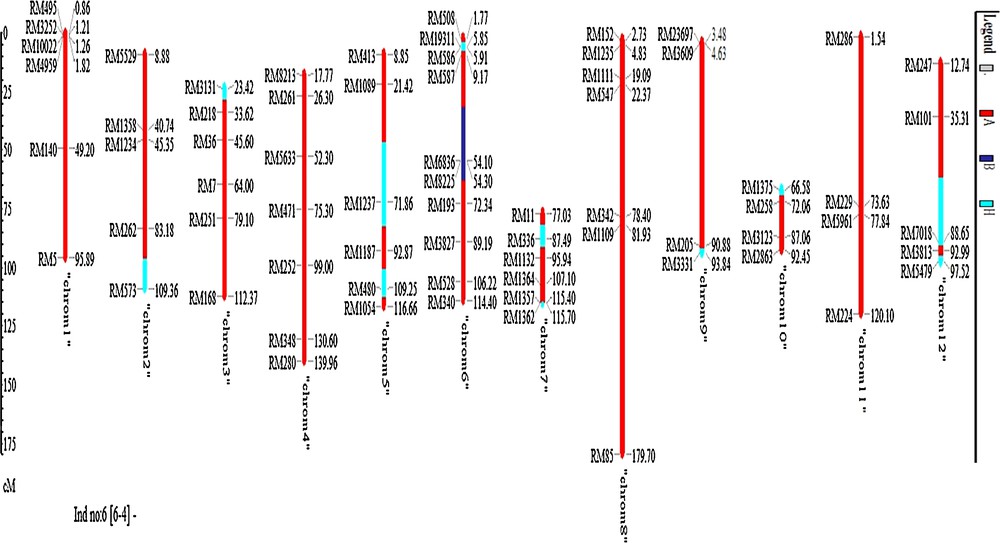
(Color online.) Chromosome-wise recurrent parent genome recovery of the best plant No. 6-4 in BC1F1 generation.
3.4 Genotyping BC2F1 generation
3.4.1 Marker-assisted foreground selection
Out of 333 plants of BC2F1 generation, in 159 plants the introgression of blast-resistant gene was confirmed using RM6836 and RM8225 markers (Fig. 5). The proportion of susceptible and resistant plants in BC2F1 generation using a foreground marker is shown in Table 3. In this study, the results fitted to the expected 1:1 ratio of BC2F1 generation (Chi2 = 0.58), which is non-significant at a probability level of 0.05. Chi2 analysis revealed that susceptible and resistant plants have a good fit to the expected testcross ratio (1:1) for a single gene model in BC1F1 and BC2F1 population. In this generation, six positive plants carrying the blast-resistant gene and having maximum morphological similarity to MR219 were selfed to produce BC2F2 seeds.
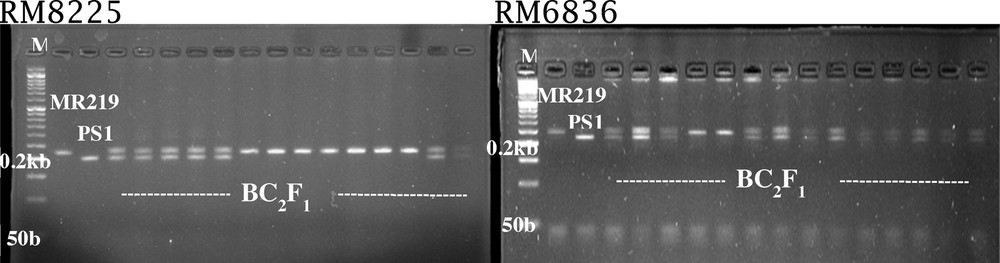
Resistant and susceptible plant screening using RM8225 and RM6836 markers in BC2F1 population.
3.4.2 Marker-assisted background selection
Background selection was performed using 70 polymorphic markers in BC2F1 plants resulting from foreground selection. The extent of recurrent parent genome (RPG) recovery ranged from 80.40% to 96.70% in BC2F1 generation (Fig. 6).
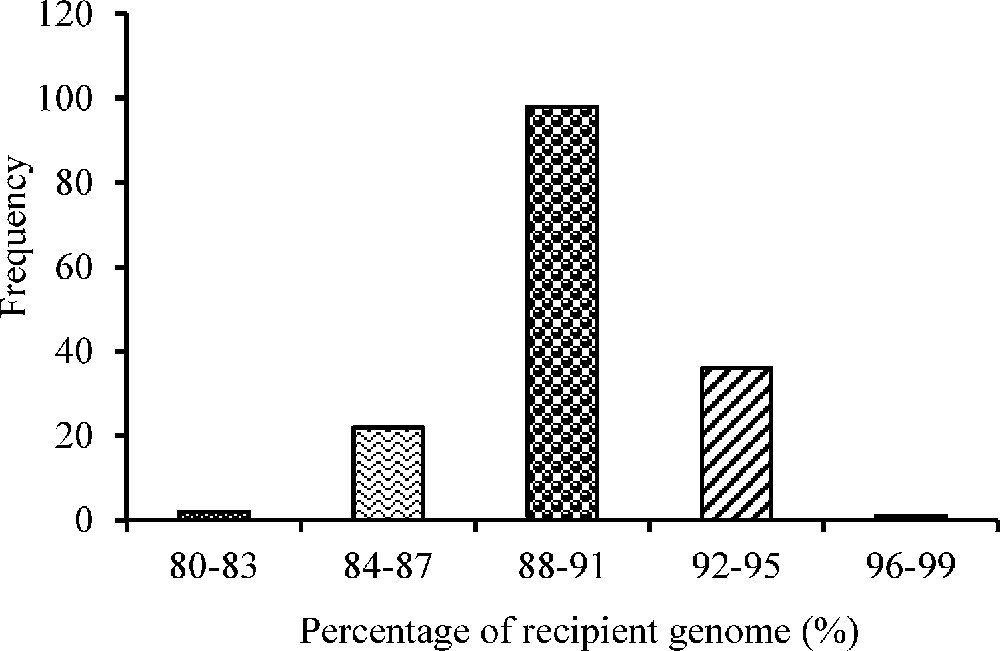
Frequency distribution of the recurrent parent genome recovery (%) in the BC2F1 population.
Six best plants (plants Nos. 6-4-1, 6-4-2, 6-4-4, 6-4-6, 6-4-7 and 6-7-13) were selected on the basis of foreground, phenotypic resemblance with recurrent parent and background information in this generation. The average recurrent parent genome recovery of the selected plants was 95.3% (Table 4). Chromosome-wise recipient allele recoveries of the selected six plants are shown in Fig. 7. For these six plants, chromosomes 1, 8 and 11 were completely recipient types. Among the six, the best plant was No. 6-4-4, and recurrent genome recoveries of this plant are shown in Fig. 8. The summary result of RPG recovery and heterozygous segment of selected individuals in BC2F1 population is presented in Table 4.
Background and introgressed segment analysis in selected lines of BC2F1 population.
| Selected individual | A (%) | B (%) | H (%) | Total (cM) | H-segments |
| 6-4-1 | 93.9 | 1.6 | 4.5 | 1162.3 | 5 |
| 6-4-2 | 95.5 | 1.6 | 2.9 | 1162.3 | 4 |
| 6-4-4 | 96.7 | 1.6 | 1.7 | 1162.3 | 5 |
| 6-4-6 | 93.8 | 1.6 | 4.7 | 1162.3 | 7 |
| 6-4-7 | 96.1 | 1.6 | 2.3 | 1162.3 | 3 |
| 6-4-13 | 96.2 | 1.6 | 2.2 | 1162.3 | 6 |
| Average | 95.3 | 1.6 | 3.05 | 1162.3 | 5.0 |
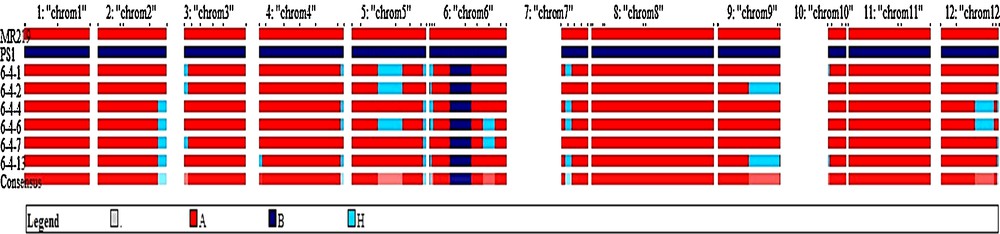
(Color online.) Analysis of recurrent parent genome recovery of selected BC2F1 plants all the 12-rice chromosomes.
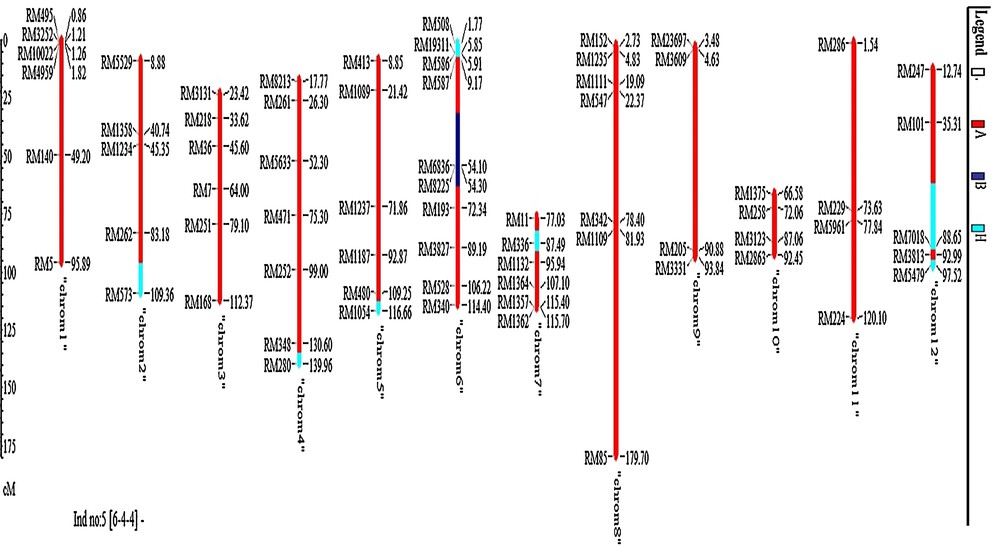
(Color online.) Chromosome-wise recurrent parent genome recovery of the best plant No. 6-4-4 in BC2F1 generation.
3.5 Genotyping BC2F2 generation
3.5.1 Marker-assisted foreground selection
In this generation, foreground selection was made to get plants appearing similar to MR219 background with homozygous resistant alleles using RM8225 and RM6836 markers. These markers showed a good fit to the expected marker segregation ratio (1:2:1) in a Mendelian fashion (single gene model), which was not significantly different (P > 0.05) (Table 5).
Marker analysis in BC2F2 population was not significantly different from the expected 1:2:1 ratio in single gene model.
| Markers | Marker segregation analysis | Chi2 (1:2:1) | Probability | ||
| AA = R | AB = SG | BB = S | |||
| RM8225 | 52 | 127 | 74 | 3.83 | 0.1473 |
| RM6836 | 54 | 132 | 67 | 1.81 | 0.4045 |
3.5.2 Marker-assisted background selection
For background selection of the advanced breeding lines (ABL), the genetic map was constructed covering 1162.3 cM with average maker distance 16.60 cM region of the O. sativa genome using 70 SSR markers. Based on background analysis with 70 polymorphic markers, the recurrent parent genome content in the selected BC2F2 lines ranged from 92.7% to 97.7%. The majority of the residual segments from the donor genome were distributed on chromosome 3, 6, 9 and 10. Perfect recovery of the recurrent parent's chromosomes was observed on chromosome 1, 2, 4, 5, 8 and 12 (Fig. 9). The percentage of substituted chromosome segments derived from Pongsu Seribu 1 in each selected improved line ranged from 1.6% to 3.8%, on the basis of the physical size of the rice chromosomes. The average proportion of the recurrent parent in the selected improved line was 95.98% (Table 6), explaining the similarity observed at the phenotypic level.
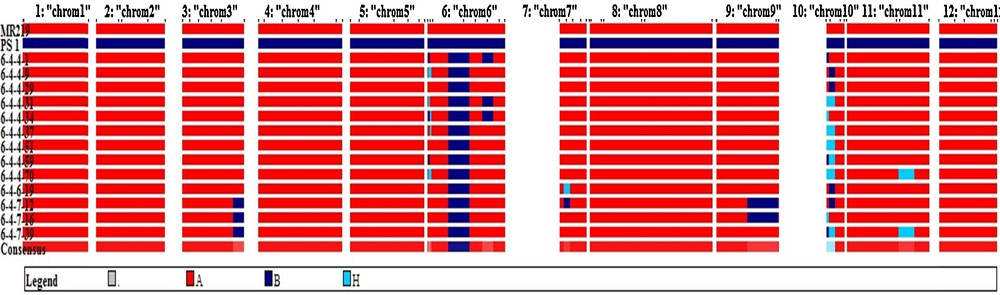
(Color online.) Recurrent parent, donor segment and heterozygous segments of the selected improved lines in MR219 background developed in this generation.
Background and introgressed segment analysis in selected lines.
| Selected individual | A (%) | B (%) | H (%) | Total (cM) | H-segments |
| 6-4-4-1 | 96.5 | 3.5 | 0 | 1162.3 | 0 |
| 6-4-4-9 | 96.9 | 2.8 | 0.3 | 1162.3 | 1 |
| 6-4-4-29 | 97.1 | 2.9 | 0 | 1162.3 | 0 |
| 6-4-4-31 | 95.2 | 3.0 | 1.8 | 1162.3 | 2 |
| 6-4-4-34 | 96.2 | 3.0 | 0.8 | 1162.3 | 2 |
| 6-4-4-37 | 96.4 | 1.6 | 2.0 | 1162.3 | 2 |
| 6-4-4-51 | 96.7 | 1.6 | 1.8 | 1162.3 | 1 |
| 6-4-4-59 | 96.7 | 2.0 | 1.3 | 1162.3 | 1 |
| 6-4-4-70 | 92.7 | 1.6 | 5.7 | 1162.3 | 2 |
| 6-4-6-19 | 96.4 | 2.9 | 0.7 | 1162.3 | 1 |
| 6-4-7-12 | 96.2 | 3.8 | 0 | 1162.3 | 0 |
| 6-4-7-16 | 97.7 | 1.8 | 0.5 | 1162.3 | 2 |
| 6-4-7-39 | 93.0 | 2.0 | 4.9 | 1162.3 | 2 |
| Average | 95.98 | 2.5 | 1.98 | 1162.3 | 1.6 |
3.6 Agro-morphological performance of the blast-resistant improved lines
Results demonstrate that a major blast-resistant Pi-gene (putative Piz) from the donor parent Pongsu Seribu 1 was successfully transferred into the MR219 genetic background and expressed similar phenotypic characteristics when compared with MR219 (Table 7), which proves that blast-resistant improved lines are similar with MR219 genetic backgrounds.
Agro-morphological trait performance of improved lines in comparison with recurrent parent MR219.
| Features | MR219 | Improved line |
| Days of 50% flowering (day) | 87.29 ± 0.56a | 89.91 ± 0.32b |
| Plant height (cm) | 96.92 ± 0.45 | 97.40 ± 0.33 |
| Effective tiller/Hill (no) | 16.04 ± 0.32 | 17.00 ± 0.40 |
| Panicle length (cm) | 25.06 ± 0.29 | 25.85 ± 0.26 |
| Number of filled grains/panicle (no) | 159.08 ± 1.28 | 161.40 ± 1.69 |
| Seed setting rate (%) | 91.25 ± 0.20 | 92.02 ± 0.25 |
| 1000 grain weight (g) | 25.37 ± 0.22 | 26.43 ± 0.14 |
| Grain length (mm) | 9.80 ± 0.01 | 9.81 ± 0.02 |
| Grain width (mm) | 2.03 ± 0.01 | 2.05 ± 0.00 |
4 Discussion
Marker screening is very essential before starting marker-assisted backcross breeding. Monomorphic markers cannot distinguish the two parental genotypes, so this type of marker bears no value in selection work. Markers that showed polymorphism were used in each backcross generation. The percentage of polymorphic marker in this study coincide more with Linh et al. [24], who screened out 477 SSR markers and found 89 (18.7%) markers polymorphism between the parents BT7 and FL478. The frequency of polymorphic SSR markers between the two parents AS996/FL478 was 12.6% [25]. In order to identify polymorphic markers for background selection, out of 309 STMS markers, 54 markers (17.47%) were found polymorphic between parents PRR78 and Pusa1460 [8]. A total of 15.1% SSR markers were found to have polymorphism between the parents Bac Thom 7 and IR64 [26].
Markers allow a good idea of how much of the recurrent parent genome recovered in any particular BC progeny and to select for the best backcross progeny available in any generation. This ability to select for recurrent parent genotype outside of the target locus can greatly reduce the number of generations required developing lines that possess the desired gene. The selection response for background analysis on non-carrier chromosomes depends on several factors, including the extent of saturation of the molecular marker map, the availability of technical resources at a given point of time, and the required levels of line conversion [27]. Reduction of the donor genome content other than the target gene requires DNA markers that are distributed evenly throughout the genome [14,19,28,29].
Background selection has already been shown to be efficient by both theoretical studies and experimental results [30,31]. As demonstrated by Tanksley et al. [32] with computer simulation, the use of molecular markers for background selection can accelerate the recovery of the recurrent parent genome by two to three generations. The number of backcross generations used in cultivar development programs may differ and depend upon a number of factors including breeder preference, genetic distance between the recurrent and non-recurrent parents, and preliminary phenotypic or genotypic selections that may occur in early backcross generations. Background selection is most efficiently performed using evenly spaced markers. A spacing of approximately 20 cM appears to be sufficient [19]. Interestingly, the findings of Visscher [33] indicated that a single marker in the middle of a chromosome may be more informative than two markers located near the ends of the chromosomes. Important factors that determine the efficiency of an MAB program are the number of target genes to be transferred, the marker map, the crossing scheme, and the applied selection strategy [34]. The genomic segments and the manner of the interaction contributing towards heterosis characterization are important to recover the original genome as far as possible, to maintain the level of heterosis of the original parent.
The marker and the gene are close together on the same chromosome (tightly linked), tend to be transmitted together in each generation. Marker-assisted foreground selection was proposed by Tanksley [35] and investigated in the context of introgression of resistance genes [36]. The use of markers for foreground selection makes the transfer of target genes feasible and economical. Marker-assisted foreground selection is mainly used for the conformation of the target alleles in the progeny generated either by selfing of a cross between donor and recurrent parent or their F1 crossed to the donor parent to determine whether an individual is kept or discarded [37]. In BC1F1 generation, RM8225 and RM6836 markers showed heterozygous plants for blast-resistant gene. After that, these two markers were used for foreground selection in the next generation. The markers RM8225 and RM6836 are linked to blast-resistant Piz gene [38,39]. The markers RM6836 and RM8225, which were found to be tightly linked with blast-resistant gene (putative Piz) in this research, were situated on chromosome 6. Piz is a disease resistance gene that has been effectively used to combat a broad-spectrum of races of the rice blast fungus M. oryzae [38]. The expected 1:1 ratio in BC1F1 generation with a Chi2 value of 0.48 was found by Mondal et al. [16]. A similar result was found in the study of Iftekharudaula [40], who also found a 1:1 ratio at BC1F1 generation with a non-significant Chi2 value of 0.28 at a probability level of 0.05.
The concept of graphical genotypes to estimate parental origin genome recovery throughout the genome was introduced [41]. RPG recoveries from 80.00% to 89.01% were found [42], which are similar to the present findings in BC1F1 generation. The present findings are more or less similar to those of Prigge et al. [43], who mentioned that mean RPG recovery in BC1 generation (computer simulated: 86% in case of Swarna and 89% in Samba Mashuri; observed: 83% in case of Swarna and 85% in Samba Mashuri) and in BC2 (computer simulated 97% in case of Swarna and Samba Mashuri; observed 95% in case of Swarna and in Samba Mashuri) generation. From the first-backcross generation (BC1), one individual of the most desirable type can be selected. If more than one individual satisfies the strongest condition, selection between them can be performed on the basis of analysis of other marker loci (located either on the carrier or non-carrier chromosomes) to determine the most desirable individual for producing BC2 [29,30,32]. If none of the BC1 individuals carries the target allele, then the backcross program failed in BC1 [44]. Several authors discussed the minimum sample size required to obtain at least one most desirable individual for producing BC2 [29,30,45].
From the background analysis, the RPG recovery was found up to 91.6% in BC2F1 generation for pyramiding blast resistance genes Piz5 and Pi54 into an elite Basmati rice restorer line ‘PRR78’ using marker-assisted stepwise backcross breeding [18]. The mean RPG recovery in BC2 generation using computer simulated 97% in case of Swarna and Samba Mashuri; observed 95% in case of Swarna and Samba Mashuri [43]. The maximum percentage of recipient alleles was found to be 93.75% and the minimum percentage of recipient alleles was 89.7% in BC2F1 generation [42], which is more similar to the present findings. In BC2F1 populations, the genetic background recovery was found up to 89.8% [26]. In an earlier study, a single-donor parent was used for the incorporation of two genes Xa13 and Xa21 into PRR78 [8]. The proportion of recurrent parent genome in the top-ten families ranged from 87.01 to 92.81% in BC2F1 generation, which was increased in BC2F5 plants where recurrent parent genome recovery ranged from 92.81 to 97.30%. In this study, the recurrent parent genome content in the selected BC2F2 lines ranged from 92.7% to 97.7%. The average proportion of the recurrent parent in he selected improved line was 95.98%. The additional recovery of RPG in the advanced generation is attributed to the fixation of the recurrent parent allele from the heterozygous alleles. The higher recovery of the RPG of this study was mainly due to the contribution of recurrent parent alleles from the backcross derivatives, which was made possible through stringent phenotypic selections. Phenotypic selection reduced both the cost and the time involved in the recovery of RPG. The application of marker-assisted background selection and its potentiality have been widely discussed, especially on the specific issues, i.e. background selection on carrier chromosome or recombinant selection, background selection on the whole genome [8,46,47], and background selection with trait of interest [48].
The presence of target trait genes must be confirmed by phenotyping in the resulting cross generation mostly at the individual level, and individual phenotypic performance is a good indicator of the genotype only if genes play a major effect on phenotypic performance, and the error of phenotyping is minimal [49]. Phenotypic selection for ‘good agronomic type’ has always been practiced along with backcross selection [50]. Conversely, the genotypic selection proposed by Young and Tanksley [28], and later referred to as background selection [30], consists in monitoring the parental origin of alleles using markers throughout the genome. Foreground selection for the trait of interest followed by stringent phenotypic selection provides superior yields [51]. The high-RPG recovery can be attributed to the stringent phenotypic selections made in every generation of backcrossing and selfing. This approach has not only reduced the time but also economized the resources involved. The phenotypic selection along with marker-assisted selection increases the recurrent parent genome and phenome recovery [52]. It is evident from the results that phenotypic selection for agro-morphological traits of MR219 coupled with marker-assisted background analysis led to hastening the recurrent parent genome and phenome recovery. The use of MAS for background selection can be very effective when the number of polymorphic markers included in the study is large, but with only 20% SSR polymorphism between the parental lines, MAS for background selection is still a distant dream for resource-limited laboratories [18]. In that case, stringent phenotypic selection for the RPP recovery is a very feasible option for the maximization of RPG recovery [53,54].
It can be concluded that marker-assisted foreground selection was successfully combined with phenotypic selection to recover yield and its components traits to develop an improved version of MR219 with inbuilt resistance to blast. There was a great acceleration of recipient genome recovery in the present study. The general conclusion is that a few well-placed markers provide adequate coverage of the genome in backcross programs. The improved lines carrying blast-resistant gene (putative Piz) in the background of MR219 were either on par or superior in agronomic performance compared to the recurrent parent MR219, with an additional advantage of blast resistance. These blast-resistant improved lines are capable of matching the productivity of MR219. The improved lines will be valuable as donors for blast resistance, and also useful for pyramiding blast resistance genes in rice breeding programs. On the basis of the present finding, it may be concluded that marker-assisted backcrossing is a potential tool to recover the recurrent parent genome in rice breeding.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgments
The authors would like to acknowledge Long-term Research Grant Scheme (LRGS), Food Security Project, Ministry of Education, Malaysia, for the financial support on rice breeding.


