1 Introduction
Rice (Oryza sativa) is one of the most important cereal crop and staple diet throughout the world. It is grown everywhere, except in Antarctica, and has great economic value. The calorie intake from rice consumed by the world population is more than 23%. The necessity for rice production has increased due to the increasing world population [1]. The rapid growth of world population, the scarcity of cultivated land and water, all these factors made blast a serious issue. The average yield potential of rice is 10 tons/ha, but farmers only harvest about 5 tons/ha [2], and this yield loss is due to insect pest and diseases.
Among biotic stresses, blast is the most threatening disease for the productivity of rice, at a time when rice production has to be increased by more than 40% by 2030 to meet the demand of people [3]. This target can be achieved by developing highly resistant rice varieties against biotic and abiotic stresses [4].
Rice blast caused by Magnaporthe oryzae is a destructing disease of rice that poses serious threats to the world's food security [5]. About 50 species of grass family are infected by M. grisea, including rice, wheat, and barley. The blast disease was first observed in India during 1919, with estimated yield losses of 4% [6], but yield losses were increased afterwards as losses only in Eastern India were estimated at around 50% [2], whereas in Japan, the crop losses varied from 20 to 100% due to various pathogens of blast disease [3]. The annual losses caused by blast disease in rice production can fulfil the annual requirement of 60 million people [7]. Rice diseases are the cause of yield loss up to 5% every year [8]. The use of resistant rice varieties has overcome the blast disease problem partially, as the pathogen have genetic variability depending on the conducive conditions. Hence, rice breeders are under a lot of pressure for developing durable resistant rice varieties.
Recently, various techniques have been used to control blast disease, including chemical and biological method, but these measures are not long lasting. Secondly, the use of fungicide is not only costly, but also hazardous to the environment. Introgression of resistant genes into rice cultivars is a very effective and safe option for controlling blast disease. Genetic studies related to blast resistance started a long time ago [9], but few cultivars have been developed with durable resistance [10] mainly due to the pathogen's variability towards virulence. Therefore, breeders recently focussed on combining genes with the help of molecular markers to get partial and complete resistance against blast [11]. The pyramiding of resistant genes or providing them with multigenic resistance is an efficient way to manage blast disease. Accordingly, different molecular techniques such as molecular mapping, gene cloning, QTL analysis, map-based cloning, marker-assisted selection have been introduced to analyse the blast-resistant genes in the rice genome [12].
The combination of a biotechnological approach and of molecular breeding can provide a high degree of resistance against M. oryzae. At present, the blast is spreading in major rice-growing areas of the world; therefore, there is an urgent need for rice varieties having high and durable resistance against blast to assure food security in the developing countries. Considering the importance of resistance development among rice cultivars against blast, this review highlights the current advances made in identifying the genes responsible for blast resistance in rice.
2 Molecular mapping of blast resistance genes
A complete rice genetic map was constructed by crossing a japonica variety (Nipponbare) with an indica variety (Kasalath) to get a population of 186 F2 individuals [13]. Sasaki [14] was the first person who described the blast disease resistance gene, and later on Kiyosawa [15] identified and introduced the first blast resistance gene (Pi-a) into the japonica rice variety of Aichi Asahi. The construction of the rice genetic map started in 1997 with the use of Restriction Fragment Length Polymorphism (RFLP) markers [16–18]. Further saturation of RFLP mapping was done by using other DNA markers such as RAPD [19,20] and SSR markers [21].
Although the genetic linkage map of many crop species was constructed by conventional breeding using morphological markers, they are numerically few and very difficult to apply as compared to molecular markers. However, with the introduction of molecular mapping, confirming the presence or the absence of blast resistance genes and finding closely linked markers becomes effortless [22].
The molecular mapping of blast resistance genes is a most convenient and direct approach for the identification of blast resistance in the rice genome. More than 100 blast resistance genes have been identified by using this approach. In recent years, major blast resistance genes and Quantitative TRAIT Loci (QTL) linked to blast resistance have been localized through molecular mapping. Moroberekan, the traditional African variety with durable resistance against various pathotypes of M. oryzae, has been used as a donor for blast resistance in many breeding programmes. By using RFLP markers, Wang et al. [23] mapped Pi5(t), Pi7(t) blast resistance genes and nine QTLs linked with partial resistance in Moroberekan variety on different chromosomes.
The complete genome of rice has been sequenced by the International Rice Genome Sequencing Project (IRGSP) in 2005 [24]; it is helpful for identifying and locating a specific position of blast resistance genes through fine mapping. More than 30 blast resistance genes [Pita-2, Pi39(t), and Pi20(t), Pi5(t), Pi15, PiCO39(t), Pi38, PBR, Pb1, Pi-kh, Pi1, Pik-m, Pik, Pik-p, Pik-s, Pi62(t), Pi157, Pit, Pi27(t), Pish, Pid1(t), Pig(t), Piy(t), Piy2(t), Pi39(t), Pi10, Pi40(t), Piz, Pigm(t), Pi33] have been identified through the molecular map-based approach. Accordingly, many blast resistance genes have been mapped on different chromosomes of rice using RFLP and SSR markers. After the sequencing of the whole rice genome, it becomes convenient to find the chromosomal location of genes linked with blast resistance through linkage to molecular markers.
The identification of molecular markers lying closely to blast resistance genes can be used for the indirect selection of genes called gene tagging [25]. Sharma et al. [26] mapped Pi54 blast resistance gene allelic to Pi-kh in the Tetep cultivar. Tetep is a blast-resistant rice variety having a broad-spectrum resistance against different pathotypes of M. oryzae. Another two blast resistance genes, Pi38 and Pi42, were identified in Tadukan and DHR9 cultivar by using different DNA markers [27,28]. The distribution of blast resistance genes is widely clustered on chromosome 6,11 and 12. Based on the mapping of blast resistance genes on different chromosomes, the genetic dissection showed that these genes are made up of nucleotide binding site-leucine-rich repeat (NBS-LRR) domain [29].
3 QTL mapping for blast resistance in rice
QTLs are regulated by more than one gene and are responsible for different characters such as yield, resistance against biotic (diseases) and abiotic factors (drought). Single marker analysis (SMA), standard interval mapping (SIM) and composite interval mapping (CIM) are three basic methods for QTL mapping [30].
The mapping population is divided into classes on the basis of the genotype at every marker locus in SMA method and demarcates QTL if significant differences in the overall mean phenotypic score are found for every group [31]. In the case of SIM at each locus, a flanking marker is used between two marker intervals. QTL can be detected easily in this method as compared to SMA. The main drawback of SIM is the identification of sometimes false QTLs because of linked and unlinked QTLs [32]. The third approach, CIM, is often used; in this method, subsets of markers are used at both linked and unlinked QTLs. The interaction of QTL declares can be detected by Composite Interval Mapping (CIM) [32].
Knowledge about tightly linked markers increases the probability of QTL detection [33]. The mapping of QTLs for blast resistance gained in importance because of partial resistance, which is durable, polygenic, non-race specific, and controlled by QTLs [34]. The partial resistance is the most suitable blast control method because it is stable and remains for a long time in comparison to complete and incomplete resistance [35,36].
The QTLs strategies have been widely used for mapping major and minor genes linked with partial resistance in rice [37]. QTL associated with blast resistance was first identified in Moroberekan cultivar grown widely in Africa [23]. There are two types of resistance: true resistance and field resistance. True resistance is associated with major gene control, qualitative and quantitative characters controlled by minor genes [38]. Most of the QTLs associated with qualitative genes have been widely seen. Sometimes, on linkage maps, the regions are very rich in genes, providing resistance to more than one pathogen because quantitative and qualitative genes exist on same position. Generally, partial resistance is a quantitative resistance governed by multiple loci called QTLs in a specific cultivar [39]. Partial resistance reduces the development of disease and reproduction of pathogens when pathogen and plant are incompatible and cannot interact [35].
From different studies, it has been proven that partial resistance can be specific [40]. Four partial resistance genes Pi34 [36,41], Pif [42], pi21 [43] and Pb1 [44] have been identified and considered as specific.
More than 350 QTLs conferring leaf blast resistance have been mapped on different chromosomes [45]. Most of these QTLs were derived from japonica indica crosses of 15 different populations. Ashkani et al. [46] found 13 QTLs, conferring partial resistance to rice leaf blast in the F3 population of local Malaysian cultivars Pongsu seribu × Mashuri. McCouch et al. [47] mapped 10 putative QTLs for blast resistance in 12 rice chromosomes, whereas Taiben et al. [48] mapped 9 QTL by using RFLP markers.
Various markers such as SSRs, RFLP and ISSR have been used to detect QTLs for blast resistance in different population and environments [49]. The 23 blast resistance loci within the QTL regions found are Pi30(t), Pi7(t), Pi34, Pi24(t), Pitq6, Pi31(t), Pi32(t), PiGD-3(t), PiGD-1(t), Pi28(t), PiGD-2(t), Pilm2, Pitq1, Pizh, Pi29(t), Pi35(t), Pitq5, Pi25(t), pi21, Pi26(t), Pi27(t), Pi24(t), Pi25(t) [50]. The detail about the mapped blast resistance genes are listed in Table 1.
List of available mapped blast resistance genes.
| Gene | Chromosomal position | Position (bp) | Position (cM) | Donor rice variety | Method of identification | References |
| Pi24(t) | 12 | 5242654–5556378 | 20.97–22.22 | Azuenca (J) | QTL mapping | [52] |
| Pi62(t) | 12 | 2426648–18050026 | 9.7–77 | Yashiro-mochi (J), Tsuyuake | Mapped within 1.9 cM | [53] |
| Pitq6 | 12 | 5758663–7731471 | 23.0–30.92 | Tequing (I) | QTL mapping | [48] |
| Pi6(t) | 12 | 1–6725831 | 1–1.68 | Apura (I) | – | [47] |
| Pi12 | 12 | 6988220–15120464 | 27.95–60.48 | Moroberekan (J) | Linkage analysis using RFLP markers | [54] |
| Pi21(t) | 12 | 5242654–5556378 | 20.94–22.22 | Owarihatamochi (J) | – | [55] |
| Pi31(t) | 12 | 7731471–11915469 | 30.92–47.66 | IR64 (I) | QTL mapping | [37] |
| Pi32(t) | 12 | 13103039–18867450 | 52.41–75.46 | IR64 (I) | QTL mapping | [37] |
| Pi157 | 12 | 12375000-15550000 | 49.5–62.2 | Moroberekan | Mapped within 9.5 cM | [17] |
| Pita | 12 | 10603772–10609330 | 42.41–42.43 | Tadukan (I) | Cloned | [56] |
| Pita-2 | 12 | 10078620–13211331 | 40.31–52.84 | Shimokita (J) | Mapped within 4.0 cM | [57] |
| Pi19(t) | 12 | 8826555–13417088 | 35.30–53.67 | Aichi Asahi (J) | Linkage analysis to other resistance genes | [58] |
| Pi39(t) | 12 | – | – | Chubu 111 (J), | Mapped within 37 kb | [59] |
| Pi20(t) | 12 | 12875000-12950000 | 51.5–51.8 | IR24 (I) | Mapped within 0.6 cM | [60] |
| PiGD-3(t) | 12 | 13950000 | 55.8 | Sanhuangzhan 2 | QTL mapping | [61] |
| Pia | 11 | 4073024–8078510 | 1.01–2.09 | Aichi Asahi (J) | – | [62] |
| PiCO39(t) | 11 | 6304007–6888870 | 25.21–27.55 | CO39 (I) | Mapped within 1.2 cM | [62] |
| Pilm2 | 11 | 13635033–28377565 | 54.54–113.5 | Lemont | QTL mapping | [48] |
| Pi30(t) | 11 | 441392–6578785 | 1.76–26.31 | IR64 (I) | QTL mapping | [37] |
| Pi7(t) | 11 | 17850000-21075000 | 71.4–84.3 | RIL29 (Moroberekan) | QTL mapping | [23] |
| Pi34 | 11 | 19423000–19490000 | 77.69–77.96 | Chubu32 (J) | QTL mapping | [36] |
| Pi38 | 11 | 19137900–21979485 | 76.55–87.91 | Tadukan (I) | Mapped within 20 cM | [27] |
| PBR | 11 | 20125000-30075000 | 80.5–120.3 | St. No. 1 | Mapped within 22.9 cM | [44] |
| Pb1 | 11 | 21425000-22850000 | 85.7–91.4 | Modan | Mapped within 12.4 cM | [63] |
| Pi44(t) | 11 | 22850000-29475000 | 91.4–117.9 | RIL29 (Moroberekan) | – | [64] |
| Pik-h | 11 | 24761902–24762922 | 99.0–99.05 | Tetep | Mapped within 1.2 cM | [26] |
| Pi1 | 11 | 26498854–28374448 | 105.99–113.49 | LAC23 (J) | Mapped within 11.4 cM | [65] |
| Pik-m | 11 | 27314916–27532928 | 109.25–110.13 | Tsuyuake (J) | Mapped within 0.3 cM | [66] |
| Pi18(t) | 11 | 26796917–28376959 | 107.18–113.50 | Suweon365 (J) | Mapped using RFLP markers | [67] |
| Pik | 11 | 27314916–27532928 | 109.25–110.13 | Kusabue (I) | Mapped within 1.4 cM | [68] |
| Pik-p | 11 | 27314916–27532928 | 109.25–110.14 | HR22 (I) | Mapped within 2.8 cM | [56] |
| Pik-s | 11 | 27314916–27532929 | 109.25–110.15 | Shin 2 (J) | Mapped within 2.7 cM | [69] |
| Pik-g | 11 | 27314916–27532930 | 109.25–110.16 | GA20 (J) | Linkage analysis to other resistance genes | [70] |
| Pise1 | 11 | 5740642–16730739 | 22.96–66.92 | Sensho | Linkage analysis using phenotypic marker | [71] |
| Pi f | 11 | 24695583–28462103 | 98.78–113.84 | Chugoku 31-1 (St. No. 1) | QTL mapping | [72] |
| Mpiz | 11 | 4073024–16730739 | 16.29–66.92 | Zenith (J) | Linkage analysis using phenotypic markers | [73] |
| Pikur2 | 11 | 2840211–18372685 | 11.36–73.49 | Kuroka (J) | Linkage analysis using phenotypic markers | [74] |
| Piisi | 11 | 2840211–19029573 | 11.36–76.11 | Imochi Shirazu (J) | Linkage analysis using phenotypic markers | [71] |
| Pi28(t) | 10 | 19565132–22667948 | 78.26–90.67 | IR64 (I) | QTL mapping | [37] |
| Pii2(t) | 9 | 1022662–7222779 | 4.09–28.89 | Azucena | Linkage analysis using phenotypic markers | [75] |
| Pi5(t) | 9 | 7825000-8250000 | 31.3–33.0 | RIL125, RIL249, RIL260(Moroberekan) | Mapped within 170 kb | [76] |
| Pi3(t) | 9 | 7825000-8250001 | 31.3–33.1 | Kan-Tao | Linkage analysis using RFLP markers | [17] |
| Pi15 | 9 | 9641358–9685993 | 38.56–38.74 | GA25 (J) | Mapped within 0.7 cM | [70] |
| Pii | 9 | 2291804–28431560 | 9.16–113.72 | Ishikari Shiroke (J) | Linkage analysis using phenotypic markers | [77] |
| Pi36 | 8 | 2870061–2884353 | 11.48–11.53 | Q61 (I) | Cloned | [59] |
| Pi33 | 8 | 5915858–6152906 | 23.66–24.61 | IR64 (I) | Mapped within 1.6 cM | [78] |
| Pizh | 8 | 4372113–21012219 | 17.48–84.04 | Zhai-Ya-Quing8 (I) | QTL mapping | [37] |
| Pi29(t) | 8 | 9664057–16241105 | 38.65–64.96 | IR64 (I) | Mapped within 0.7 cM | [37] |
| Pi17(t) | 7 | 22250443–24995083 | 89.00–99.9 | DJ 123 | Mapped within 1.8 cM | [70] |
| Pi22(t) | 6 | 4897048–6023472 | 19.5–24.09 | Suweon365 (J) | QTL mapping | [55] |
| Pi26(t) | 6 | 8751256–11676579 | 35.00–46.70 | Azucena (J) | QTL mapping | [45] |
| Pi27(t) | 6 | 5556378–744329 | 22.22–2.97 | IR64 (I) | Mapped within 21.6 cM | [37] |
| Pi40(t) | 6 | 16274830–17531111 | 65.09–70.12 | O. australiensis(W) | Mapped within 1.8 cM | [79] |
| Piz | 6 | 10155975–10517612 | 40.6–42.07 | Zenith (J) | Mapped within 0.43 cM | [67] |
| Piz-t | 6 | 14675000 | 58.7 | Toride 1 | Cloned | [56] |
| Pi9 | 6 | 10386510–10389466 | 41.5–41.55 | O. minuta (W) | Cloned | [80] |
| Pi25(t) | 6 | 18080056–19257588 | 72.32–77.03 | Gumei 2 | QTL mapping | [21] |
| Pid2 | 6 | 17159337–17163868 | 68.63–68.65 | Digu | Cloned | [81] |
| Pigm(t) | 6 | 10367751–10421545 | 41.47–41.68 | Gumei 4 | Mapped within 70 kb | [82] |
| Pi26(t) | 5 | 8751256–11676579 | 35.00–46.70 | Gumei 2 (I) | QTL mapping | [21] |
| Pi23(t) | 5 | 10755867–19175845 | 43.02–76.70 | Sweon 365 | QTL mapping | [55] |
| Pi10 | 5 | 14521809–18854305 | 58.08–75.41 | Tongil | Mapped within 6.7 cM | [83] |
| pi21 | 4 | 5242654–5556378 | 20.97–22.22 | Owarihatamochi | QTL mapping | [55] |
| Pikur1 | 4 | 24611955–33558479 | 98.44–134.23 | Kuroka (J) | Linkage analysis using phenotypic marker | [74] |
| Pi39(t) | 4 | 26850000–27050000 | 107.4–108.2 | Chubu 111 (J) | Mapped within 0.3 cM | [59] |
| Pi(t) | 4 | 2270216–3043185 | 9.08–12.17 | Tjahaja | Linkage analysis using phenotypic marker | [17] |
| Pid1(t) | 2 | 21875000–22475000 | 87.5–89.9 | Digu | Mapped within 11.8 cM | [84] |
| Pig(t) | 2 | 34346727–35135783 | 137.38–140.54 | Guangchangzhan (I) | Mapped within 11.8 cM | [85] |
| Pitq5 | 2 | 37625000–39475000 | 150.5–157.9 | Teqing | QTL mapping | [48] |
| Piy1(t) | 2 | 38300000–38525000 | 153.2–154.1 | Yanxian No. 1 | Mapped within 1.6 cM | [86] |
| Piy2(t) | 2 | 38300000–38525001 | 153.2–154.1 | Yanxian No. 1 | Mapped within 3.0 cM | [86] |
| Pib | 2 | 38300000–38525000 | 153.2–154.1 | Tohoku IL9 | Cloned | [87] |
| Pi25(t) | 2 | 34360810–37725160 | 137.44–150.90 | IR64 (I) | QTL mapping | [21] |
| Pi14(t) | 2 | 1–6725831 | 1–26.90 | Maowangu | Linkage analysis using isozyme markers | [70] |
| Pi16(t) | 2 | 1–6725831 | 1–26.91 | Aus373 (I) | Linkage analysis using isozyme markers | [88] |
| Pit | 1 | 2270216–3043185 | 9.08–12.17 | Tjahaja | Mapped within 770 kb | [56] |
| Pi27(t) | 1 | 6230045–6976491 | 24.29–27.90 | IR64 (I) | Mapped within 21.6 cM | [37] |
| Pi24(t) | 1 | 5242654–5556378 | 20.97–22.22 | Azuenca (J) | QTL mapping | [35] |
| Pitp(t) | 1 | 25135400–28667306 | 100.54–28667306 | Tetep | Co-segregation marker was identified | [89] |
| Pi35(t) | 1 | 33000000–34150000 | 132.0–136.6 | Hokkai 188 (J) | QTL mapping | [90] |
| Pi37 | 1 | 33110281–33489931 | 132.44–133.95 | St. No. 1 (J) | Cloned | [91] |
| Pish | ||||||
|
Pi67(t)
Piss2 Pise2 |
Map position still not identified | |||||
| Pise3 |
4 Marker-assisted selection of blast resistance genes
Marker-assisted selection is a promising and efficient way to select the desirable characters indirectly in a breeding program by using different markers such as SSR, ISSR, RFLP, RAPD [49]. The selection of genes of interest depends upon the location of the marker and distance lying within gene and marker. The identification of marker linked to the gene of interest is a basic prerequisite of marker-assisted selection through linkage analysis. The tagging of genes becomes possible with the advent of molecular markers responsible for resistance to different pathogens [92].
The application of marker-assisted selection is very powerful in case of blast resistance breeding because single or a few genes are involved in the resistance mechanism [32]. The development of durable blast-resistant rice varieties against the different strains of M. oryzae progressed through the availability of different molecular markers applied for marker-assisted selection [93]. A dominant marker specific for the particular blast resistance gene has been developed, it can be used in a marker-assisted selection programme for the development of an improved blast-resistant rice cultivar [94].
SSR markers are not only used in marker-assisted selection for blast resistance genes, but also in diversity analysis and inheritance studies. Three major blast resistance genes, Pi-b, Pi-k, and Pi-ta, have been extensively studied by different researchers and are molecularly characterized. The inheritances from one generation to another have enabled these genes to become suitable for marker-assisted selection because of their Co-segregation with respect to the specific markers [26,69].
The selection of blast resistance genes through marker-assisted selection is very precise because of the governing true interaction of the particular resistance (R) gene with the avirulence gene. Several blast resistance genes Pi-z, Pi-ta Pi-b, Pi-k and Pi-I have been introgressed through marker-assisted selection into susceptible varieties [95,96]. Rice breeding for blast resistance has gained an importance after the characterization and analysis of R genes because the dissection and identification of these genes within local varieties become effortless. Blast resistance genes, Pita [97], Pi37 [98], Piz [99], Pi35 [90], and Pi9 [100] have been introgressed through the application of marker-assisted selection. The number of disease-resistant cultivars can increase with the continuous transfer of newly DNA markers to breeders.
To facilitate the incorporation of R genes into elite breeding lines, few blast resistance genes have been characterized molecularly and tightly linked markers such as SSR, SNP have been developed [69,101]. MAS may increase the genetic improvement if information about the molecular markers associated with the disease resistance QTL becomes available [102]. Successful examples of MAS for blast resistance in rice are displayed in Table 3 (Fig. 1).
Some successful examples of MAS for blast resistance in rice cultivars.
| S.no | Trait | Genes | Type of markers | Method applied for transferring target gene | References |
| 1 | Blast resistance | Pi1, Piz-5 Pita | RFLP | Marker-assisted-selection applied for combining these genes into a single cultivar Co-39 | [65] |
| 2 | Blast resistance | Pi54 | SSR | Marker-assisted-selection applied to improve Pusa Basmati 1 from Tetep | [124] |
| 3 | Blast resistance | Pid1, Pib and Pita | SSR | Introduced into G46B cultivar through MAS | [84] |
| 4 | Blast resistance | Pi2 | SSR | Introduced into Zhenshan97B through MAS | [84] |
| 5 | Blast resistance | Pi1, Pi2, Pi33 | SSR | Introgressed into Jin23B cultivar through marker-assisted backcrossing | [129] |
| 6 | Blast resistance | Pi1, Piz-5, Pi2, Pita | RFLP, SSR, ISSR | Introduced into C039 cultivar through MAS | [65] |
| 7 | Blast resistance | Pi-9 (t) | pB8 (gene specific marker) | MABC applied to introgress into Luhui17 cultivar | [130] |
| 8 | Blast resistance | Pi-ta | Gene specific marker | MAS applied | [131] |
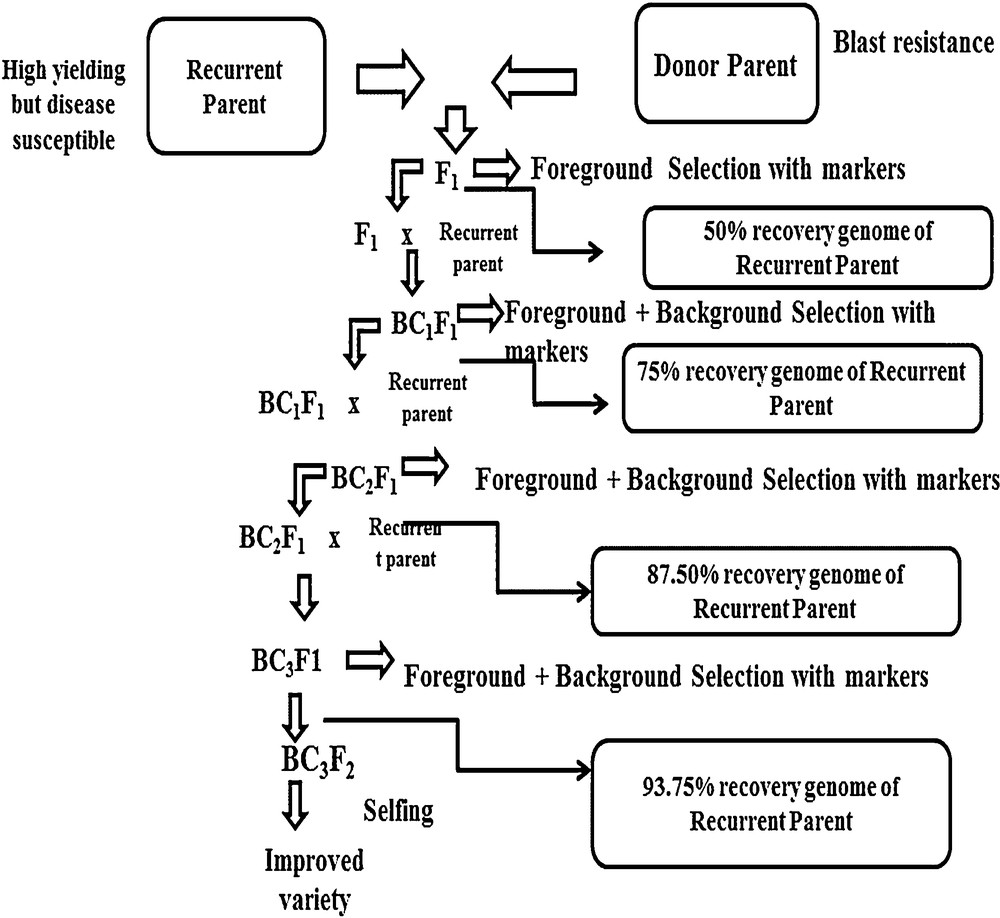
Diagrammatic representation of general steps of marker-assisted backcross breeding.
5 Identification of molecular markers linked to blast resistance genes
The identification of tightly linked markers with genes of interest is the basic step for the map-based cloning, molecular mapping, marker-assisted selection and other molecular techniques involved in biotic stress resistance in rice.
Recently, applications of molecular markers have been widely used in agriculture, especially in rice improvement research [16]. Molecular markers for blast resistance can be produced on different sites either on targeted or random sites to make more informative and dense genetic maps [103,104]. Accordingly, a large number of databases are available to select the molecular marker linked to Pi genes.
Molecular markers are powerful tools for monitoring the introgression of the gene of interest and for the detection of polymorphism within the different species [105]. The monitoring of an individual resistance gene and its pyramiding into a breeding line is very difficult; only a DNA marker tightly linked with the gene provides the straightforward way for the selection of multiple blast resistance genes. The selection of the desired traits can be made at an early stage with the advent of molecular markers [106]. When a molecular marker is found very close to the gene of interest, it acts as a tag to be used for indirect selection in the molecular breeding program [25].
After the discovery of molecular markers, the selection of target traits becomes easier and many new cultivars have been developed accordingly. Nowadays, breeders are focusing on marker-assisted selection instead of using conventional breeding because it reduces the time for phenotypic selection, saves input costs, brings more reliability to select a desired trait with no influence of environmental factors [51].
Many DNA markers are directly linked with Pi genes in rice including simple sequence repeats (SSRs), amplified fragment length polymorphisms (AFLPs), and cleaved amplified polymorphic sequences (CAPS), random amplified polymorphic DNAs (RAPDs), restriction fragment length polymorphisms (RFLPs), single-nucleotide polymorphisms (SNPs) and small insertions/deletions (InDels). SSRs and CAPS are PCR-based markers and require only a small amount of DNA for genotyping. These markers are very precise and cost effective and can be applied for the selection of plants containing blast resistance genes in rice at an early stage. Small InDels and SNPs markers are found in abundance and dispersed widely in the rice genome [107]. On the basis of information on these markers, Hayashi et al. [56] developed nine PCR-based markers linked with blast resistance in rice. These markers help in finding a gene within the desired target genome regions. Microsatellite markers, also called SSRs, are widely used for screening the blast-resistant and susceptible varieties. The difference between two varieties is based on polymorphism [105].
SSRs markers are the markers of choice for breeders because of their highly polymorphic nature; PCR-based and codominant SSR markers are widely used for studying genetic diversity among populations to estimate the proportion of the genome introgressed from the donor to the parents. More than 2000 SSR markers have been developed, linked with different blast resistance genes, which have thus accelerated the MAS and MABC for the improvement of rice cultivars [69]. The selection of allele with the advent of SSR markers is very precise, authentic and reliable because of the co-segregation of the marker. The target allele can segregate into homozygote, heterozygote and recessive allele with the application of SSR markers. The international Rice Microsatellite Initiative (IRMI) has developed a microsatellite map covering all 12 rice chromosomes, at least one microsatellite at the distance of 0.5 cM [108]. The multiline carrying more than one blast resistance gene can be developed with the help of SSRs markers, without screening against pathogens. Ashkani et al. [109] found six SSR markers, RM168, RM5961, RM413, RM1233, RM6836 and RM8225 directly associated with leaf blast resistance in a Malaysian rice variety. The major blast resistance genes Pi-b, Pi-kh and Pi-ta linked SSR markers have been developed and successfully introgressed into different susceptible rice cultivars through MAS [11,26,101,110].
Restriction Fragment Length Polymorphism (RFLP) is a non-PCR-based technique used to construct the genetic map of a species. The selection of the desired DNA sequence depends on different probes obtained after digestion with restriction enzymes. RFLPs markers are codominant; informative, reliable but very costly and time consuming. Several RFLPs markers linked with blast resistance genes conferring complete resistance have been mapped on different chromosomes in rice. A RFLP marker linked with Pi-2(t) was identified by Yu et al. [111], whereas Tohme et al. [112] also identified a RFLP linked with Pi-4(t). The inheritance of complex traits such as blast resistance becomes possible with the advent of RFLP [113]. Several SNP, STS, InDel, and CAPS markers have been identified, linked with different blast resistance genes and are mentioned in Table 2.
Blast resistance genes and their respective tightly linked DNA markers.
| Type of marker | |||||||
| Gene | Chromosome | SSR | SNP | STS | InDel | CAP | References |
| Pi27(t) | 1 | RM151,RM259 | [114] | ||||
| Pi37 | 1 | RM302,RM212 FPSM1,FPSM2,FPSM4 | [98] | ||||
| Pi35(t) | 1 | RM1216,RM1003 | [90] | ||||
| Pitp(t) | 1 | RM246 | [89] | ||||
| Pid1(t) | 2 | RM262 | [84] | ||||
| Pig(t) | 2 | RM166,RM208 | [114] | ||||
| Piy1 | 2 | RM3248,RM20 | [86] | ||||
| Piy2 | 2 | RM20,RM3248 | [86] | ||||
| Pib | 2 | RM138,RM166,RM208,RM266,RM138,RM166 | b213,b28,b2, b3989,Pibdom | [86] | |||
| pi21 | 4 | P702D03 | OPF62700 OPF62700 | [84] | |||
| Pi39 | 4 | RM3843,RM5473 | [115] | ||||
| Pi10 | 5 | OPF62700 | [116] | ||||
| Pi40(t) | 6 | RM3330,RM527 | S2539 | [79] | |||
| Piz | 6 | z60510,z5765,z56592,z565962 | [56] | ||||
| Piz-t | 6 | z60510,z5765,zt56591 zt5659 | z4794 | [56] | |||
| Pigm(t) | 6 | z4794 | C26348 | [82] | |||
| Pi36 | 8 | RM5647 | S47656 | CRG2,CRG3, CRG4 | [61] | ||
| Pi33 | 8 | RM72,RM44 | [78] | ||||
| Pi5(t) | 9 | JJ817 | 94A20r,76B14f 40N23r |
[117] | |||
| Pia | 11 | yca72 | [76] | ||||
| PiCO39 | 11 | RGA8, RZ141 yca72, RGACO39 |
[62] | ||||
| Pi38 | 11 | RM206,RM21 | [27] | ||||
| Pik | 11 | K6438, K6415K6415,K8823,K39512 | K6816 K2167 |
[56] | |||
| Pik-m | 11 | K6816 K2167 |
[56] | ||||
| Pik-p | 11 | [56] | |||||
| Pi-kh | 11 | RM206,TRS26,TRS33,RM144,RM224,RM1233,RM224 | [118] | ||||
| Pik-s | 11 | RM144,RM224RM1233RM144,RM224 | [69] | ||||
| Pita | 12 | OSM89,RM155,RM7102RM7102 | ta642,ta801ta3, ta577,ta5,Pi-ta 440,Pita-1042,Pi-ta 403 | [56] | |||
| Pita-2 | 12 | ta801,ta642, ta3 ta577,ta5 | [56] | ||||
| Pi20(t) | 12 | RM1337,RM5364,RM7102 | [119] | ||||
| Pi39(t) | 12 | 39M6, 39M7 | [59] |
6 Marker-assisted backcrossing for blast resistance
Marker-assisted backcross breeding has received attention for the introgression of blast resistance genes into the genomic background of the susceptible varieties of rice. The main objective of marker-assisted backcross breeding is to reduce the donor genome content into the target varieties.
Mainly, two steps i.e., foreground selection and background selection, are applied in backcrossing. Foreground selection is done by selecting a specific marker near the target genome, whereas background selection is applied by dispersing markers throughout the genome for the recovery of the recurrent parent [120].
Backcross breeding has been widely used for the introgression of qualitative characters such as resistance against specific pathotype disease controlled by a single dominant gene [121]. The main purpose of MABC is to transfer the desired character along with recovering the recurrent parent characters. Marker-assisted backcrossing is currently playing an important role in plant breeding and genetic engineering for the development of blast-resistant cultivars [122]. The implementation of marker-assisted backcross breeding is time saving and cost effective over conventional plant breeding [123].
Recently, blast resistance genes Piz5 and Pi54 have been introgressed into the genetic background of PRR78 rice variety from donor parent C101A51 and Tetep, and blast-resistant lines have been developed with the application of marker-assisted backcross breeding [124]. The selection was based on foreground markers RM287 and RM206 by following repetitive backcrossing. Pi1 leaf blast resistance gene has been introgressed into D521 line derived from donor line BL122 [125].
With the application of marker-assisted backcrossing elite, 304 parental lines of hybrid rice have also been improved with bacterial blight and blast resistance [126]. Marker-assisted backcross breeding has been applied for both biotic and abiotic stresses. Recently from IR64 cultivar submergence, tolerant gene Sub1 has been introgressed into OM1490 variety [127]. The QTL saltol derived from a salt-tolerant variety has also been introgressed into popular cultivars of Vietnam [128]. The successful example of MAS is mentioned in Table 3 and general steps of marker assisted backcross breeding have been described in Fig 1.
7 Cloning of blast resistance genes
This is the most effective and direct approach for the characterization and identification of the blast resistance gene. The first step for cloning is the fine mapping of genes closely linked with the DNA marker, followed by molecular cloning of the desired gene.
So far, more than 100 blast resistance genes have been identified in both indica and japonica rice cultivars. Out of them, only 19 blast resistance genes have been cloned and characterized molecularly [5]. Map-based cloning is also called positional cloning; it sometimes reverses genetics because it does not require information about the gene or its gene product; however, the knowledge of the chromosomal location of the desired gene is essential [132].
The interest in cloning and characterizing blast resistance genes among researchers increased when Wang et al. [87] successfully cloned Pi-b gene from cultivar Tohoku IL9. Pi-ta was another major blast resistance gene cloned after Pi-b [110]. After the successful cloning of these genes, Sharma et al. [26] isolated and characterized the Pi-kh gene from cultivar Tetep. Afterwards Pi-d2, Pi9, Pi-2, Piz-t, Pi36, Pi37, Pik-m, Pi5, Pit, Pid3, Pi21, Pis-h, PB1, Pi-k, Pik-p, Pia have been cloned from different indica and japonica cultivars (see Table 4 for references).
List of cloned blast resistance gene.
| S.no | Name of gene | Year of cloning | Chromosome | Cultivar name | Type of rice (I – indica; J – japonica) | Characteristics | References |
| 1 | Pib | 1999 | 2 | Tohoku IL9 | J | NBS–LRR | [87] |
| 2 | pi-ta | 2000 | 12 | Tadukan | I | NBS–LRR | [110] |
| 3 | pi54 | 2005 | 11 | Tetep | I | NBS–LRR | [26] |
| 4 | pi2 | 2006 | 6 | Jefferson | J | Lectin receptor | [133] |
| 5 | pi-zt | 2006 | 6 | Zenith | I | NBS–LRR | [134] |
| 6 | pi9 | 2006 | 6 | 75-1-127 (101141) | Oryza minuta | NBS–LRR | [80] |
| 7 | pid2 | 2006 | 6 | Digu | I | NBS–LRR | [81] |
| 8 | pi36 | 2007 | 8 | Q61 | I | CC–NBS–LRR | [61] |
| 9 | pik-m | 2008 | 1 | Tsuyuake | I | NBS–LRR | [135] |
| 10 | pi37 | 2008 | 11 | St. No. 1 | J | NBS–LRR | [98] |
| 11 | pi21 | 2009 | 9 | Owarihatamochi | J | CC–NBS–LRR | [44] |
| 12 | pid3 | 2009 | 1 | Digu | I | CC–NBS–LRR | [136] |
| 13 | pit | 2009 | 6 | Tjahaja | J | NBS–LRR | [56] |
| 14 | pi5 | 2009 | 4 | Moroberekan | J | NBS–LRR | [117] |
| 15 | pb1 | 2010 | 1 | Modan | I | CC–NBS–LRR | [137] |
| 16 | pish | 2010 | 11 | Shin 2 | J | CC–NBS–LRR | [51] |
| 17 | pik | 2011 | 11 | Koshiminori | I | CC–NBS–LRR | [138] |
| 18 | pikp | 2011 | 11 | HR22 | I | CC–NBS–LRR | [56] |
| 19 | pia | 2011 | 11 | Aichi Asahi | J | CC–NBS–LRR | [139] |
The genetic dissection of cloned genes revealed that they contain the NBS-LRR domain, except Pid-2, which contains the receptor kinase domain. The maximum distributions of cloned genes have been found on chromosomes 6, 11 and 12 (Fig. 2).
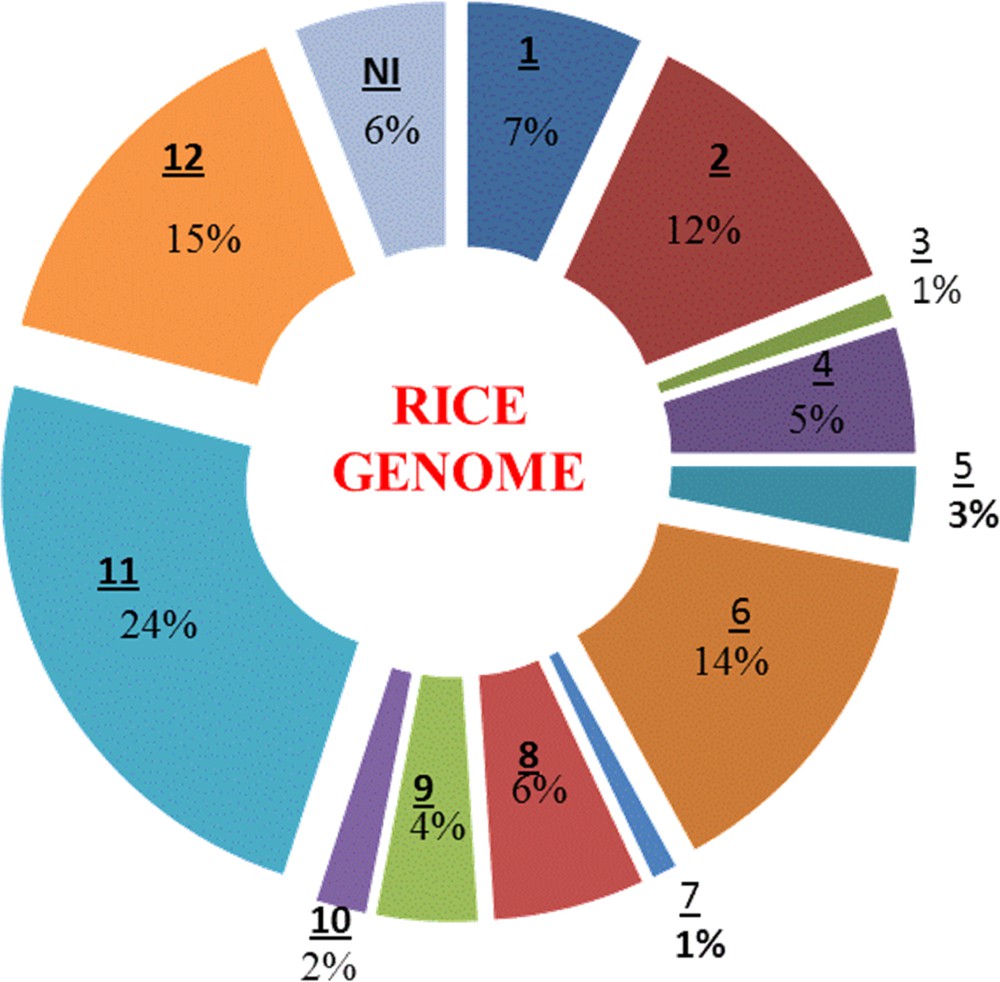
(Colour online.) Distribution of blast resistance genes among 12 chromosomes of rice. Underlined numbers represent the chromosome position. NI indicates that no information available.
The figure was modified from Sharma et al. [26].
NBS-LRR is one of major protein family, providing disease resistance. NBS-LRR is further divided into two subfamilies based on the presence of Toll/interleukin-1 receptor (TIR) or coiled-coil (CC) motifs in the amino-terminal domain [140]. Among 19 cloned blast resistance genes, 10 genes fall into a category of NBS-LRR proteins, eight into CC-NBS-LRR type domain, while Pid-2 possesses a unique type of protein called B-lectin receptor a having serine threonine kinase gene (Fig. 3).
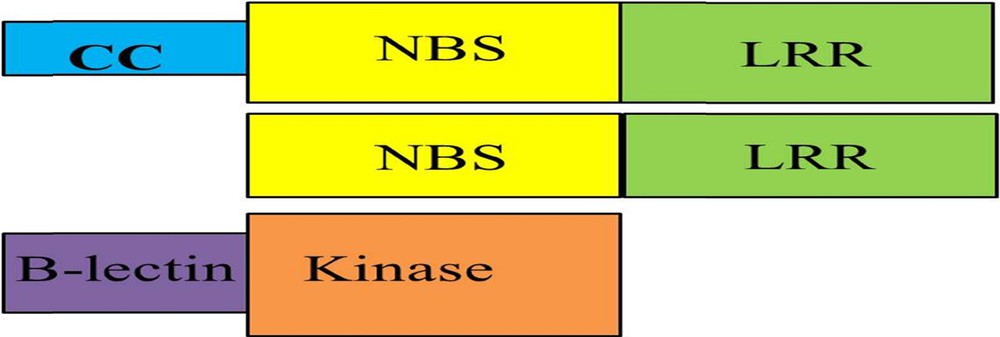
(Colour online.) Different types of domain combinations found among blast resistance genes present in rice.
8 Allele mining strategies for blast resistance genes
The major goal of plant breeding is to find the superior/novel agronomic characters. The amount of variation found within the plant germplasm contributes significantly to achieve this goal. All the plants have the potential to collect a beneficial allele for certain agronomic traits, whereas the evolution of novel or new alleles depends heavily upon the natural mutations [141]. There are many factors involved in mutation in plants such as tranvertions, InDels, and transitions. Allele mining is an advanced and newly developed technique to determine the allelic variation at candidate gene regulating important agronomic traits. With the passage of time, allele mining opened a new avenue for the validation of blast resistance genes and mining of favourable alleles present at Pi loci [142]. The identification of alleles of identified gene/locus responsible for controlling any given trait or identification of superior allele from the natural population is called allele mining. Usually two approaches are used for allele mining in the naturally developed populations, i.e. Targeting Induced Local Lesions in Genomes (TILLING) [143], called EcoTilling and Re-sequencing [144], or sequencing-based allele mining. In TILLING, heteroduplex analysis is carried out to directly identify the induced point mutation in the gene. TILLING enables the identification of single-base-pair (bp) allelic variation in a target gene in a high throughput manner. This technique employs the mismatch-specific endonuclease in order to detect induced or natural DNA polymorphisms [145]. The fragments produced are separated and the site of mutation can be identified by the fragment size analysis. Sequence- and sequencing-based allele mining consists in a PCR-based amplification of alleles of a gene in varied genotypes, then in DNA sequencing to recognize the nucleotide variance in alleles. The sequences are further analysed for the presence of SNPs, and InDels. The SNPs and InDels help to construct the haplotyps. Haplotypes are the basis for understanding the effect of mutation on organization and gene structure. Both above-described approaches have been widely used for the dissection of important blast resistance genes from wild rice species. From the identified rice blast resistance genes, Pi1 [146], Pik [147] Pi-ta [148], Pi1 [111], Pi2 [149], Pib [87], Pi9 [133], Pi5 [117], and Pik-p [150] are considered as very well defined. The identification of blast resistance genes Pi9, Pi2 and Piz-t from different rice cultivars showed that these genes tend to be an allele. The physical locations of these genes are on the same locus, but express different levels of resistance spectra in different cultivars [106,151,152]. Allele mining for Pi-kh, Pi-ta and Pi-zt genes has been also reported in Indian land races of rice [153]. Allele mining can be essential for determining and utilizing novel alleles, hidden in genetic diversity. The identified blast resistance genes through allele mining strategy have been described in Table 5.
Current status of allele mining for blast resistance in rice.
| Name of gene | Trait control | Allelic gene | Rice variety | References |
| Pid3 | Blast resistance | Pid3-A4 | Digu | [154] |
| Pi-ta | Blast resistance | Pita (Konibora) | (Konibora) | [155] |
| Pi-kh | Blast resistance | Pi54 | Tetep | [26] |
| Pi-2 | Blast resistance | Pi2-2 | Jefferson | [156] |
9 Conclusion
The demand for rice is increasing day by day due to the increase in the population globally. To fulfil the dietary requirements, high yielding varieties of rice are required. The high yield can be obtained by reducing the yield losses due to pest and disease damage. To overcome these losses because of diseases, molecular breeding techniques have expedited the rice research at the DNA level to explore the mechanism of host–pathogen interaction. Rice breeding has gained many direct and indirect advantages from rice genomic research, but further research is still required to exploit tools, knowledge, and research in breeding programs.
The comparison between the sequences of different blast resistance genes with different spectrum of resistance will lead to identify the specific DNA region responsible for the resistance specificity. The application of molecular techniques to produce the broad-spectrum resistance in rice is increasing consistently to reduce the cost of inputs.
Although many blast resistance genes have been mapped and cloned, they provide resistance to specific pathotypes. Due to the genetic stability and pathogenic variability, the rice varieties have rapidly been broken down. The need for mapping and cloning has increased so as to identify more specific and suitable blast resistance genes that confer resistance to different pathotypes of M. oryzae.
The QTL identification, fine mapping, marker-assisted selection, cloning of blast resistance genes are valuable tools to understand the genetic mechanism and function of genes. The QTL mapping requires a large population; however, it is not possible to identify the precise position of the desired genes until cloning and fine mapping is done. Hundreds of QTLs has been cloned that control different traits in rice, but still more QTLs needs to be cloned and identified in the future. Map-based cloning methods have investigated many QTLs for specific resistance genes, which can be applied in rice breeding programmes.
This article will be helpful to study the current advanced methods of gene identification that could be utilized to study the structure, function and regulation of genes involved for providing resistance against blast disease. The different molecular techniques will accelerate the development of resistant cultivars to meet the demand for rice at the global level.
10 Future prospects
The majority of world countries consume rice as a basic food because of its high carbohydrate value and also because it is financially affordable. Rice production methods have been improved in the last decades due to rice breeding and improved cultural practice, but blast diseases have caused heavy loses to rice and are considered as major constraints to sustainable rice production. The instability of pathogens in the field subjected rice breeders to pressure for producing more resistant varieties having durable resistance.
There will be more blast resistance genes introduced in the future by dissecting the interaction between the R genes and AVR protein. To date, few blast resistance genes have been characterized; in the future, more blast resistance genes will be characterized.
A differential system for gene characterization is necessary. Currently, monogenic lines and nearly isogenic lines have been released with the collaborative research project of IRRI-Japan. These lines will be useful as gene sources/donor parents for developing blast-resistant varieties and international standard differential varieties used for the characterization of blast resistance genes. Breeders have to explore more selection markers applicable for MAS on the basis of an increasing number of blast resistance genes.
Till to date, blast genes identified by different researchers provide resistance against the leaf blast. Most of the blast-resistant rice varieties produced yet provide monogenic resistance; sometimes, because of monogenic resistance, the cultivar loses its resistance. The tagging blast resistance genes with molecular markers have already been achieved. It will be more feasible if breeders work with several genes and pyramid into elite rice cultivars. The IR64 variety is the best example: it contains at least five blast resistance genes that provide them with durable resistance.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgments
The authors would like to acknowledge Long-Term Research Grant Scheme (LRGS), Food Security Project, Ministry of Education, Malaysia, for the financial support for conducting research on rice breeding. The author would also like to acknowledge Sindh Agriculture University Tandojam, Sindh, Pakistan, for providing financial support.


