1 Meiosis and recombination: a general overview
Meiosis is essential for the fertility of most sexually reproducing eukaryotes. This peculiar cell division halves the number of chromosome sets, compensating the chromosome doubling occurring during fertilization. Meiosis is thus a key step in the sexual life cycle. It consists in two rounds of chromosome segregation that follow a single round of DNA replication (Fig. 1). In a vast majority of species, the first round of chromosome segregation separates the homologous chromosomes – and is at the origin of the ploidy reduction – while the second round of chromosome segregation separates the sister chromatids – as during a mitotic division (Fig. 1).
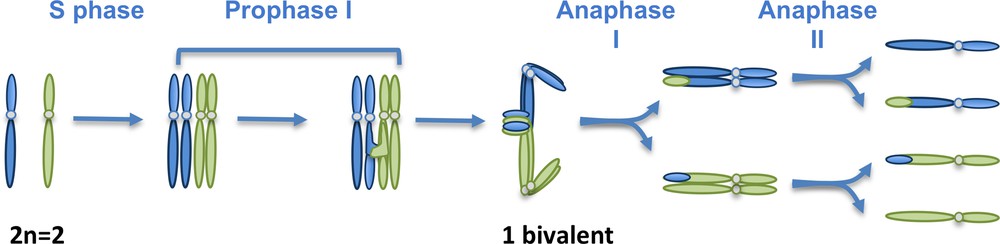
Schematic representation of a meiotic division. In meiocytes, as in any sporophytic cell, a set of chromosomes of maternal origin (blue) coexists with a set of chromosomes of paternal origin (green). They correspond to pairs of homologous chromosomes (or homologs). Here, a hypothetical organism with a diploid number of chromosomes of 2 has been chosen. Replication (S phase) duplicates each chromosome into two sister chromatids that are kept together by the action of cohesins (not shown). Meiosis consists in the succession of two rounds of chromosome segregation (Anaphases I and II) after a single S phase. During prophase I, homologous chromosomes recombine and associate into bivalents. Meiosis I separates the homologous chromosomes, while meiosis II separates the sister chromatids.
The success of ploidy reduction that occurs during the first meiotic division depends on the preliminary association of the homologous chromosomes into pairs forming a structure called bivalent. In most species, bivalent formation relies on meiotic recombination, and more precisely on the occurrence of crossovers (COs). COs correspond to large reciprocal exchange of two non-sister chromatids that connect physically the two homologous chromosomes (Fig. 2). In these species, the presence of at least one CO per bivalent is an absolute requirement for the proper segregation of the homologous chromosomes and thus to obtain fully viable gametes.
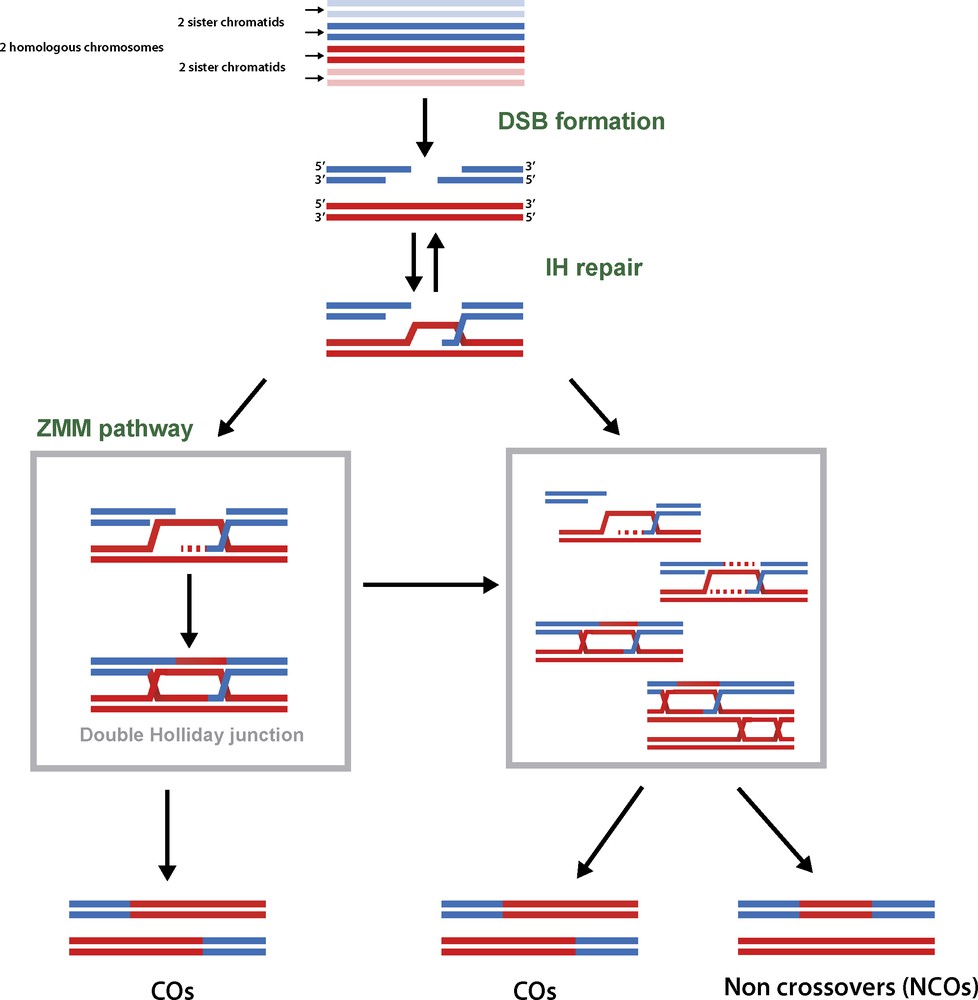
Overview of the meiotic recombination mechanisms. Meiotic recombination is initiated by the induction of large amount of DNA double strand breaks (DSBs). The 5′ extremities of these DSBs are resected to generate 3′ single stranded DNA molecules that will eventually be involved in homology search and intact DNA duplex invasion. After invasion, the recombination intermediates can be stabilised by the ZMM proteins, and channelled toward the formation of double Holliday junctions, a subset of which will eventually be resolved as COs. Alternatively, joint molecules will be submitted to anti-CO activities and repaired into NCOs. A small fraction of these can also generate COs.
Meiosis is a very important source of genetic variability. First because COs reorganise allelic combination within chromosomes and second because anaphase I reshuffles parental chromosomes into daughter cells. Over the last twenty years, considerable progresses have been made in deciphering the mechanisms that govern meiotic recombination. The aim of this review is not to get into the details of such mechanisms (for which a number of recent reviews will be provided), but more to discuss the implication of these discoveries.
2 Meiotic recombination specificities
Recombination corresponds to one of the molecular processes available to repair the DNA lesions that affect the two strands of a DNA molecule – the double strand breaks (DSBs). It is encountered in all three branches of life – bacteria, archaea and eukaryotes – and relies on the selection of a similar or identical DNA matrix as a template to repair the compromised one. It is therefore described as a conservative DNA repair process in opposition to the mechanisms such as non-homologous end joining (NHEJ) where the repaired DNA duplex looses genetic information [1]. Homologous recombination generates two types of products: the COs that correspond to reciprocal exchanges of large chromosomal fragments between homologous chromosomes and the non-crossovers (NCOs) that correspond to a local repair of the DNA with a short and non-reciprocal replacement of one DNA sequence with a homologous one (Fig. 2). The different steps and overall mechanisms that govern meiotic and somatic recombination are very similar, but meiotic recombination shows some interesting specificities in regards to its somatic counterpart.
2.1 The initiation of meiotic recombination
Contrary to somatic recombination, the DNA lesions that initiate meiotic recombination are not fortuitous but genetically programmed [2]. Besides, an intriguing feature of meiotic recombination is that in most organisms, the number of DSBs induced at meiosis is surprisingly high, estimated in many species to several hundreds [2,3]. Meiotic recombination therefore starts by submitting the gamete mother cells to an outstanding level of DNA damage. This is particularly unexpected for cells on which relies the transmission of the genetic heritage. As expected, multiple layers of controls that regulate DSB localization, rate and fate have been characterised [4].
A number of meiotic proteins required for meiotic DSB formation have been identified, mostly in Saccharomyces cerevisiae, Schizosaccharomyces pombe, Arabidopsis thaliana and Mus musculus [2]. SPO11 is one of the few of these “DSB proteins” to be widely conserved among species. In the late 1990s, it was found that SPO11 shows similarity with the catalytic domain of a topoisomerase form Archaea – the topoisomerase VI or TopoVI – and that it could be purified covalently linked to the 5′ extremities of the meiotic DNA DSBs [5,6]. Since then, it is assumed that SPO11 carries the catalytic activity responsible for DSB formation. However, no evidence of such biochemical activity could be obtained, and hardly any progress in the understanding of the function of the other DSB proteins has been done. However, the recent characterisation in A. thaliana of the long-sought partner of SPO11, corresponding to the complementary part of the topoisomerase from Archaea [7], as well as the identification of distantly related homologues in vertebrates, insects and ascomycetes [8] now makes us revisit the nature, structure and functions of the meiotic DSB forming complex [9].
2.2 The choice of the template for DSB repair
Homologous recombination requires the search, the recognition and the use of a “similar” DNA molecule as a matrix for DNA repair. In a diploid cell after replication either the sister chromatid or the two chromatids of the homologous chromosomes are available as a matrix for repair of the damaged DNA duplex (Fig. 2). During meiosis, the repair is biased toward the homologous chromosome at the expense of the sister chromatid – a process called inter-homologous bias. This is an absolute prerequisite for the formation of COs between homologous chromosomes and requires the involvement of a meiotic specific machinery that prevents the use of the sister chromatids for repair [10]. How this could be achieved so successfully is still very much astonishing. The scale at which such search is undertaken appears to be the 3′ single stranded DNA generated after DSB processing (Fig. 2) and involves the eukaryotic homologues of the bacterial RecA proteins, RAD51 and DMC1 [10]. Larger scale mechanisms such as chromosome and nuclei movements, telomere attachment to the nuclear envelope and telomere clustering (also called the “bouquet”) could also be involved, but no understanding of these long time described peculiar meiotic chromosome dynamics is yet available. Other important layers of control are brought by the structural components of the meiotic chromosomes. Meiotic chromosomes are organised as chromatin loops emanating from a protein axis. During prophase, the two protein axis of the homologous chromosomes become linked along their entire length by the polymerisation of the central element of the synaptonemal complex (SC) that physically connect the homologous chromosomes. It has become clear from these last year studies that most of the biochemistry of recombination is tightly linked with the SC components. This is particularly the case concerning the step of template choice since many components of the chromosome axis have been shown to regulate the efficiency of inter-homologous repair [11].
In polyploid organisms, the situation is even more complex since possible DNA templates available for repair must be extended to the chromatids of the homoeologous chromosomes. The use of the homoeologous chromosomes for repair would be very detrimental to the cells because it generates chromosome rearrangements such as deletion and translocation. The mechanisms that prevent homoeologous and favour homologous repair are still completely unknown and represent a major issue to be solved in the future [12].
2.3 During meiosis the outcome of recombination is compelled toward CO formation, but the number of COs is nevertheless strongly constrained
As already stated, homologous recombination can generate two types of molecular products, either COs or NCOs. During somatic DNA repair, CO formation is largely avoided and several biochemical pathways that promote NCO formation have been characterised [13]. This can be related to the fact that COs in somatic cells can have dramatic consequences such as loss of heterozygocity or chromosomal rearrangements. One major discovery of the last decade in the meiotic recombination field is the identification of a pro-CO machinery specific to meiosis, the “ZMM pathway” [14]. The proteins of this pathway – Zip1, Zip2/SHOC1, Zip3 and/or Hei10, Zip4, Mer3, Msh4 and Msh5 – plus Mlh1/Mlh3 (which are not classified as ZMM but are part of the same pathway) are present in the vast majority of the eukaryotes studied so far, with the notable exception of S. pombe. Disruption of the ZMM pathway drastically reduces CO formation, and it is generally estimated that it controls the formation of 50–100% of the COs, according to species. Immunocytological studies performed in Sordaria macrospora, A. thaliana, tomato, and mammals revealed that ZMMs are active on numerous early recombination intermediates, but that only a few of these will eventually mature into COs [11]. A major challenge for the future will be to understand the mechanisms controlling CO designation out of these ZMM-marked intermediates.
It has recently been revealed that, acting antagonistically to the ZMM pathway, several somatic-inherited anti-CO pathways are active during meiosis [15]. Disruption of these anti-CO machineries in A. thaliana and in S. pombe leads to unleash CO number [16,17]. It therefore appears that, in regards to somatic DNA repair, the meiotic recombination machinery has acquired the capacity to protect a subset of recombination intermediates from the anti-CO activities acting during recombination. It is nevertheless surprising to observe that the average number of CO per bivalent is poorly variable, always above 1 (illustrating the already mentioned “obligatory CO”) but in the vast majority of the cases below 3 and this whatever is the physical size of the chromosome [12]. It remains to be understood what are the reasons for such constrained number of COs.
2.4 Where does recombination occur?
When the study of meiosis became widespread at the beginning of the 20th century, it became quickly evident that meiotic recombination rate and localization were not homogenous throughout the genomes. Along the chromosomes of almost all species, domains with high recombination rates alternate with domains where recombination rates are significantly lower than the genome-wide average. In budding yeast, Arabidopsis, wheat, and humans, more than 80% of the recombination events occur in less than a quarter of the genome [18]. The understanding of the factors that profile the CO landscape is still very fragmented, but it appears that the shape of the CO distribution is the result of a series of controls that could be interconnected and could vary from species to species: DSB positioning in correlation with chromatin marks, the repair fate of each DSB (CO, NCO or inter-sister repair), the structure of the chromosome axis and the meiotic chromosome dynamics. The recent identification in A. thaliana of the first meiotic mutants that modify the localisation of meiotic recombination events without changing the average level of their occurrence opens new and promising roads of investigations. First, PSS1, a Kinesin1-like protein, has been shown to be required for wild-type CO location since its disruption leads to the loss of the obligatory CO [19]. Kinesins being component of the chromosome mobility machinery, it provides the first evidence of the importance of meiotic chromosome dynamics in the regulation of CO localisation. Second, disruption of AXR1, a key component of the neddylation machinery, provokes the clustering of COs in the sub-telomeric regions of the chromosomes, without affecting the average CO number [20]. Neddylation is an important post-translational regulatory mechanism, common to most eukaryotes and linked to targeted protein degradation. The deciphering of the mode of action of this pathway during recombination will undoubtedly provide information on the general regulatory mechanisms implemented during recombination and resulting in a given CO landscape.
3 Concluding remarks
The actual knowledge on meiotic recombination mechanisms suggests that meiotic recombination has evolved from an ancestral somatic DNA repair process, forcing the system toward CO formation. This appears to be correlated with the selection of strongly deleterious processes, such as DNA double strand break formation and inter-homologs COs promotion, despite the strong risks for chromosomal rearrangements that these processes are known to provoke. Interestingly, recombination-free meiosis also exists, for example in the male from Drosophila melanogaster or the female from Bombyx mori [21], but it is strikingly poorly represented among species, and in any cases, never observed for the two sexes of a single species. Since recombination generates new combinations of alleles but also destroys favourable associations, it is likely that the prevalence of sex with limited number of recombination events corresponds to an optimised evolutionary situation, in terms of stability but also adaptability.
Disclosure of interest
The author declares that he has no competing interest.


