1 Introduction: a myriad of sex determination mechanisms
Sexual reproduction is an extraordinarily widespread phenomenon that assures the production of new genetic combinations in nearly all eukaryotic lineages. Although the core mechanisms of sexual reproduction (meiosis and gamete fusion or syngamy) are highly conserved, the pathways that determine the male and female specificities are remarkably diverse across eukaryotes and appear to have had rapid turnover rates during evolution [1].
Genetic determination of sex is mediated by sex chromosomes, whose key feature is the sex-determining region (SDR) that carries the sex-determining factor(s) and usually does not recombine in the heterogametic sex. The SDR can be as small as a single locus or as large as an entire chromosome. Sex chromosomes are derived from autosomes and have independently evolved many times in different eukaryotic lineages. Sex chromosomes can arise in species with separate sexes in which sex is determined by environmental cues (such as in turtles, where temperature determines the sex of developing embryos) and can also arise in hermaphrodites (that is, individuals with both male and female sex organs). Sex chromosomes are subject to unique evolutionary forces including sex-specific selection, asymmetrical sheltering of deleterious mutations, hemizygosity, or dosage compensation [2]. Besides sex determination, sex chromosomes play a prominent role in several evolutionary processes such as speciation and adaptation [3].
Traditionally, most of our knowledge on the biology and evolution of sex chromosomes stems from a few well-studied model organisms, notably mammals, birds, Drosophila and the plant Silene latifolia. Animals and plants, however, represent just two out of the 21 major groups of Eukaryotes (Fig. 1A) and the universality of the mechanisms driving the evolution of the sexes is difficult to assess without a broader view that takes into account the scope of the tree of life. In this context, the brown algae, a group of complex multicellular eukaryotes, are extraordinary comparative models to look at how the sexes have originated and evolved in a lineage that has been independently evolving from animals and plants for more than a billion years.
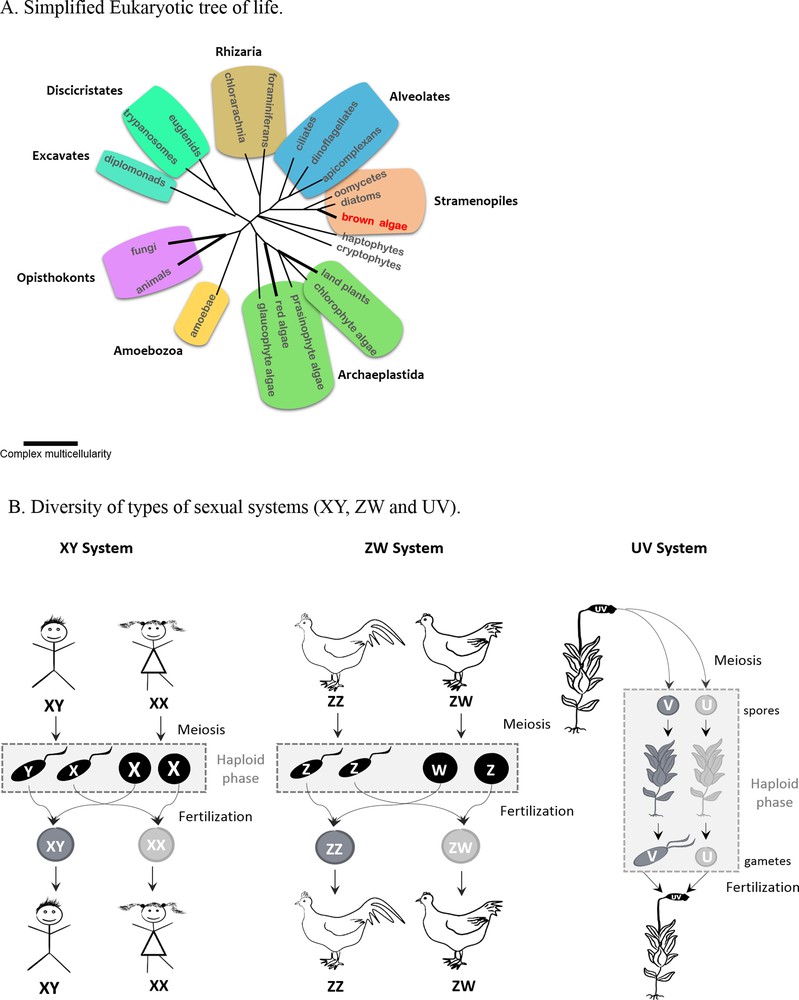
A. Simplified Eukaryotic tree of life, adapted from [24]. B. Diversity of types of sexual systems (XY, ZW and UV). In organisms with diploid life cycles, sex is determined in the diploid phase of the life cycle, after fertilization. In XY systems, the sex of the embryo depends on the chromosome carried by the sperm cell, X or Y. In ZW it is the female egg that determines the sex of the individual. In organisms such as some algae and mosses, that alternate between gametophyte and sporophyte generations (haploid-diploid life cycles), sex is expressed during the haploid (gametophyte) phase of the life cycle. The sexual system in this case is called UV systems. In contrast to XY and ZW systems, sex in UV systems depends on whether the spores receive a U or a V chromosome after meiosis (not at the fertilisation stage).
Moreover, while studies so far have focused on diploid sex determination systems (the classical XY or ZW systems, Fig. 1B), haploid phase sex-determination systems (UV systems such as those of mosses and algae) [4] have been less studied. Contrasting the properties of XY/ZW and UV chromosomes, together with comparisons with species that lack sex chromosomes entirely, provides an outstanding chance to assess the relative importance of the forces driving the evolution of each system.
2 Evolution of sex chromosomes: new insights from UV systems
In recent years, comparative studies of sex chromosomes of different ages from both animals and plants have led to the emergence of a model for the evolution of sex chromosomes [5]. A typical sex chromosome pair derives from a pair of autosomes through the acquisition of genes involved in sex determination. If more than one locus involved in sex determination is located on the chromosome, recombination between loci is suppressed, leading to the establishment of a non-recombining region on the nascent sex chromosome, the sex-determining region or SDR. The formation of this non-recombining region has important consequences for the evolution of this part of the genome: repetitive DNA tends to accumulate, leading to an increase in SDR size and degeneration of genes within the non-recombining region. At a later stage, deletion of non-functional DNA from within the SDR may lead to a decrease in the physical size of the SDR. The evolutionary processes driving the expansion of the SDRs (and corresponding shrinkage of the pseudoautosomal region, PAR) have been the subject of research, mostly theoretical, for many years and the currently accepted view is that this process is driven by the recruitment of genes with differential selective benefits for the two sexes (sexually antagonistic genes) into the SDR [6], to resolve sexual conflicts.
The evolution of sex chromosomes is largely impacted by the genetic mechanism of sex determination. In organisms where sex is expressed in the diploid phase, such as most animals and land plants, one sex is heterogametic (XY or ZW) whilst the other is homogametic (XX or ZZ). In these systems only the Y or W contain non-recombining regions because the X and Z recombine in the homogametic sex. In contrast, in some algae and bryophytes the male and female sexes are genetically determined after meiosis, during the haploid phase of the life cycle (Fig. 1B). This type of UV sexual system exhibits specific evolutionary and genetic properties that have no exact equivalent in diploid systems. A number of verbal predictions regarding UV evolution had been formulated, but they have long lacked empirical support. The U and the V have been predicted to show similar characteristics because they function in different individuals and both are non-recombining; furthermore, degeneration should be modest because purifying selection is expected to act on the SDR genes during the haploid phase of the life cycle, and the U and V are expected to experience mainly gene gain [7].
Recent work using the model brown alga Ectocarpus (Box 1) has made an important contribution to the empirical validation of predictions in relation to sex chromosomes. The Ectocarpus UV system displays some striking similarities with XY and ZW systems, such as low gene density and accumulation of repeated DNA in the non-recombining, sex-determining region. This highlights the universality of some of the processes shaping sex chromosome evolution across distant lineages. The Ectocarpus system has, however, had a distinct evolutionary trajectory. Remarkably, although this UV system is very old (> 100 Myr), the SDR has remained small [8,9]. A possible explanation for this feature was suggested by the low number of sex-biased genes, implying that sexual conflict in Ectocarpus may be insufficient to drive extensive SDR expansion [8,10]. This result is therefore consistent with the view that sex antagonism may drive SDR expansion. Surprisingly, both the male and female SDR haplotypes showed signs of degeneration despite the action of purifying selection during the haploid phase of the life cycle, which is expected to purge mildly deleterious mutations and therefore prevent degeneration.
Taken together, we can say that studies using the SDR of “exotic” organisms have increased our knowledge on the evolutionary trajectory of UV systems, and have provided, and will certainly continue to provide, a broader phylogenetic dimension to sex chromosome evolutionary models.
While the genomic and evolutionary characteristics of the sex-specific regions are increasingly well understood, the recombining regions (the ‘pseudoautosomal region’ or PAR) of the sex chromosomes have been less studied. The PARs are expected to behave like autosomes and recombine during meiosis; therefore genes in this region are inherited in an autosomal rather than a strictly sex-linked fashion. Genomic-scale analyses have been carried out for the PARs of a less than a handful of old XY or ZW sexual systems [11]. Recent work using the well assembled PAR of the UV system of Ectocarpus showed that although PARs recombine at a rate that is not different from any other region of its genome, this region exhibits a number of structural and evolutionary features that are typically associated with regions of suppressed recombination [10]. The PAR of this UV system has accumulated genes that are differentially expressed during the sporophyte versus gametophyte generation of the life cycle, compared to the rest of the genome, and these generation-specific genes exhibited clear signs of accelerated evolution. Generation-antagonistic forces may therefore affect the evolution of the PARs [10]. This, and the work described above on the sex-determining region of the Ectocarpus UV sex chromosomes, has brought to light the critical impact of the life history of the organisms, in particular the type of life cycles, in the evolution of sex chromosomes.
3 Beyond Ectocarpus: brown algae as models to understand the origin and evolution of sexual systems
In this context, the brown algae are emerging as interesting models for investigating the origins and evolution of sex chromosomes because they exhibit a bewildering variety of life history traits, e.g. reproductive systems, types of life cycle and sex chromosome systems in a single group, and there appears to have been considerable switching between variants of these different features on an evolutionary timescale. The maintenance of this range of variability in a single group is actually quite unique among the eukaryotes, and clearly points to a complex evolutionary history of the underlying sex-determination systems.
The brown algae represent a unique group for studies of the evolution of the sexes, particularly with regard to the evolution of gamete size sexual dimorphism as this group exhibits a high level of variability for this trait, ranging from isogamous, through anisogamous, to oogamous systems [12]. Anisogamy has evolved several times in this group and somewhat surprisingly, oogamy seems to be the ancestral state in brown algae [13]. This opens the intriguing possibility that oogamy may evolve towards isogamy, despite the fact that transitions from oogamy towards isogamy are difficult to explain from a theoretical point of view [14]. The diversity of levels of gamete dimorphism and the multiple transitions between gamy levels within the brown algae are highly interesting in the context of recent views on the evolution of gamete size dimorphism and its consequences for the evolution of the SDRs. Evidence has been reported for linkage between the mating type (MT) locus and a gene(s) controlling gamete size in the green alga Chlamydomonas, and it has been suggested that the MT locus in a related oogamous species underwent significant expansion correlated with the evolution of gamete size sexual dimorphism [15]. Yet, the molecular basis of gamete size control and its link to sex determination remains unclear. The brown algae, thus, may represent great comparative systems to complement work done in the green lineage.
Differences in gamete size in anisogamous and oogamous brown algal species may influence other reproductive characteristics. In particular, gamete size may be one of the factors that determine whether a gamete is capable of undergoing asexual reproduction through parthenogenesis should it fail to encounter a gamete of the opposite sex. In isogamous, anisogamous and oogamous brown algal species there are marked differences between the relative parthenogenetic capacities of male and female gametes [13,16]. A trend is apparent: usually both male and female gametes of isogamous brown algal species are capable of parthenogenesis whereas only the female gametes of anisogamous species are parthenogenetic (i.e. in the latter parthenogenesis is a sexually dimorphic trait). Neither the male nor the female gametes undergo parthenogenesis in oogamous species. Overall, these tendencies suggest that gamete size may influence parthenogenetic capacity up to a point, but that in oogamous species the large female gamete is specialised for zygote production and is no longer capable of initiating parthenogenetic development [12].
The brown algae are the Eukaryotic lineage that exhibits the broadest diversity of types of life cycle and a widest range of different sexual systems. For example, sexuality is expressed during the diploid phase of the life cycle in brown algae with diploid life cycles (dioecy) such as the fucoids, whereas it is the haploid gametophyte generation that exhibits sexuality (dioicy) in algae such as Ectocarpus that have haploid-diploid life cycles [12]. The selective pressures leading to the evolution of these different systems are distinct: whilst dioecy might evolve from monoecy to limit inbreeding (due, in the latter, to the fertilisation of female gametes by male gametes produced by the same organism), this is unlikely to be the case for dioicy because deleterious mutations should be efficiently purged during the extensive haploid phase of the life cycle. Similarly, genetic sex determination is expected to operate differently, with XY or ZW systems occurring in dioecious species but UV systems occurring in dioicous species. When the different types of brown algal life cycle are mapped onto a phylogenetic tree, the distribution pattern suggests that there has been considerable switching between different life cycle strategies and sex chromosome systems during the evolution of this group [13,16], and that dioicy was the ancestral state in the brown algae, and the transition to dioecy presumably required an intermediate state of co-sexuality (e.g., monoecy) with epigenetic sex differentiation (as opposed to genetic sex determination). Phylogenetic comparative methods will be valuable to fully characterise and reveal correlations between these life cycle and reproductive traits and their relationships with the evolution of sex chromosomes across the brown algal lineage.
4 Master sex-determining genes across lineages
Sex chromosomes harbour the master-switch gene that determines gender. A few master sex determination genes have been identified across the eukaryotes [17]. The transcription factor DMRT1 is involved in sex determination in deeply divergent taxa such as Drosophila, C. elegans, medaka fish and frogs. High-mobility group (HMG)-domain proteins have been implicated in gender determination in both vertebrates and fungi, and the mating type (MT) locus of the green alga Volvox also contains a HMG-domain gene [18]. Remarkably, a HMG-domain gene has been identified in the SDR of Ectocarpus and this gene represents therefore a strong candidate for the sex-determining gene in this species. If this gene acts as the master sex determination gene in Ectocarpus, which is very distantly related to previously identified species with HMG sex-determining genes (more than a billion years of independent evolutionary history), this will raise very important questions about the evolution of sex-determination gene networks across the Eukaryotes, suggesting shared or convergent mechanisms in brown algae, fungi and animals.
Disclosure of interest
The authors declare that they have no competing interest.
Acknowledgements
Work in the Algal Genetics group is supported by the CNRS, the ANR (project Bi-cycle ANR-10-BLAN-1727 and project Idealg ANR-10-BTBR-04-01), the Interreg program France (Channel)-England (project Marinexus), the UPMC and the European Research Council (grant agreement 638240). LM had a PhD grant (ARED) from the Région Bretagne.


