1 Introduction
Transposable elements (TEs) are ubiquitous components of eukaryotic genomes. The first TEs were discovered by B. McClintock decades ago [1], as “controlling elements” involved in the variegation of kernel colour in maize. At the time McClintock was awarded her Nobel Prize in 1983, TEs had been discovered in all eukaryotic organisms where they were looked for and this has since been confirmed by the discovery that TEs are by far the major constituents of plant and animal genomes [2]. TEs are classified into two categories [3]. Class-I elements, also referred to as retrotransposons, transpose via RNAs intermediates, through a copy and paste mechanism. Class-II elements, the transposons, transpose through excision and re-insertions (a cut-and-paste mechanism). Class-I elements can undergo large transpositional bursts, which lead in some cases to catastrophic genomic amplifications. This is well exemplified in human with the LINE L1 which is present at more than 100,000 copies [4], or in rice, for which we showed that the retrotranspositional activity of only three LTR-retrotransposon families led to a doubling of genome size of the wild rice species Oryza australiensis in the last 2 Myr [5]. This propensity of TEs to densely populate eukaryotic chromosomes rather helped solve one of the most striking paradox in biology, i.e. the « C-value paradox » [6], by showing that the large extent of genome size variation in eukaryotes can be accounted for mainly by TEs, while the gene content among plants and animals remains remarkably constant (except for some variations caused by polyploidization or segmental duplications). However, while solving the paradox, this discovery led to a paradigm, built on the “selfish gene” theory, which classified TEs in a non-genic and thus non-functional compartment of the genome [7]. Nevertheless, the mutagenic nature of TEs has always been a “stone in the shoe” of some evolutionary genomicists who could not reconcile this putative negative effect on fitness (caused by the inactivation of genes) with such evolutionary success. Recent studies have suggested that transposition may on the contrary be beneficial to organisms and at the origin of major biological innovations in the course of evolution. For instance, the transpositional burst of the transposon mer20 was shown to be at the origin of the placenta through the reprogramming of developmental genes [8] in mammals. More recently, Chuong et al. [9] showed that a similar process involving an endogenous retrovirus (ERV) was at the origin of innate immunity also in mammals. Interestingly, several cases of TE-mediated phenotypic changes have also been reported in plants, some of which being of agronomic interest, like the grape colour [10] the blood orange phenotype [11], or the Mantel abnormality in date palm [12].
2 The new paradox
Despite the fact that TEs are present in most genomes and exhibit polymorphisms in both natural populations and domesticated gene pools, which is an indirect evidence of their transpositional activity, the analysis of TE-driven genomic transgenerational changes in various organisms have shown that, in a vast majority, TEs are not transpositionally active. Over the past ten years, significant advances have been made in our understanding of how transposition is suppressed through epigenetic mechanisms. Transposition in plants and animals is notably limited through TE silencing ensured by epigenetic mechanisms [13,14]. These mechanisms include the deposition of epigenetic marks, such as DNA methylation and heterochromatic histone post-translational modifications. In addition, in-depth analysis of angiosperms genomes shows that they often harbour traces of more ancient transposition bursts, but these are in the form of short, deleted and degenerated copies of TE families [15], suggesting that TE-related sequences are eliminated over time. There are two main mechanisms of LTR-retrotransposon elimination: small deletions [16] and illegitimate recombinations between LTRs, leading to solo-LTR sequences and the elimination of circular molecules harbouring one LTR and the internal region of the element [17]. The combined actions of silencing and elimination should lead to complete elimination of TEs from most species, which is exactly the opposite of what is observed. This new paradox could only be solved with the discovery of the mechanisms allowing TEs to circumvent the silencing machinery of their host. We recently proposed a model positing that this may actually be achieved through horizontal transfers [18].
3 Horizontal transfers of transposable elements
HTs refer to the transmission of genetic material across reproductive barriers. They have been evidenced for TEs for many years [19]. However, until recently, horizontal transfers of TEs (HTTs) were essentially described in animals [20,21], with only very few cases in plants [22]. HTTs are usually detected using three criteria: phylogenetic incongruence between the host and TE phylogenies, patchy TE distributions in phylogenies, and/or a high similarity between TEs from distantly related species. In a pioneering large-scale comparative genomic survey [18], we showed that HTTs are in fact very frequent in flowering plants, with an estimation of more than two millions among monocots and dicots within the last two million years. We also showed that TEs were transpositionally active following their transfer, leading in some species to significant genomic amplification. We therefore proposed that HTTs should be a key mechanism of the evolution of eukaryotic genomes by allowing TEs to escape the silencing machinery of their host genome.
4 Mechanisms of horizontal transfers
The mechanisms through which genomic DNA could be horizontally transferred among species remain not fully understood, whether they concern host genes (referred to as horizontal gene transfers, HGTs) or TEs (HTTs). In any case, one could reasonably argue that the prerequisite of HTs at large is the biological proximity of the species involved in the transfer. In plants, this may be best evidenced in the case of grafting. Although the evidences of grafting-based HGTs have been so far provided experimentally [23], the occurrence of natural grafting in ecosystems may be a source of HTs. More generally, host–parasite interactions have been proposed to favour HTs. In animals, Gilbert et al. [20] identified several HTTs among species involved in parasitic relationships, while HGTs between the parasitic plant genus Cuscuta and its host Plantago [24] and between Striga and Sorghum [25] were also evidenced. Interestingly, biotic interactions may be at the origin of HTs among species infected by the same parasite. For instance, Ivancevic et al. [26] showed that the LINE BovB has been horizontally transferred to several taxonomic groups as diverse as marsupials, ruminants, squamates or monotremes, through the same arthropod vectors (ticks). Gilbert et al. [27] showed the presence of several TE-related sequences (transposons and retrotransposons) in 21 moth baculovirus populations. Several of these TEs have undergone HTs among several insect species, demonstrating the possible role of encapsidation of foreign DNA in viruses in HTs. In plants, Ghoshal et al. [28] showed that cucumber necrosis virus can encapsidate TEs from their host (Nicotiana benthamiana). These three possible routes of HTs are schematized in Fig. 1. The question of the transfer of DNA through feeding has been debated, especially in the context of the evaluation of the risks associated with the consumption of GMOs, although no experimental evidence that these may occur has ever been provided. There is however one intriguing case of inter-kingdom HGT that was recently documented: the sea slug Elysia chlorotica is able to perform photosynthesis because of the presence of chloroplasts in his epidermis. These organelles are in fact transferred from the algae Vaucheria litorea, which is part of its diet. Although this transfer is not heritable (the photosynthetic capability of the slug ends when it stops consuming algae), it represents an interesting case of transfer of a biological function from the alimentation [29].
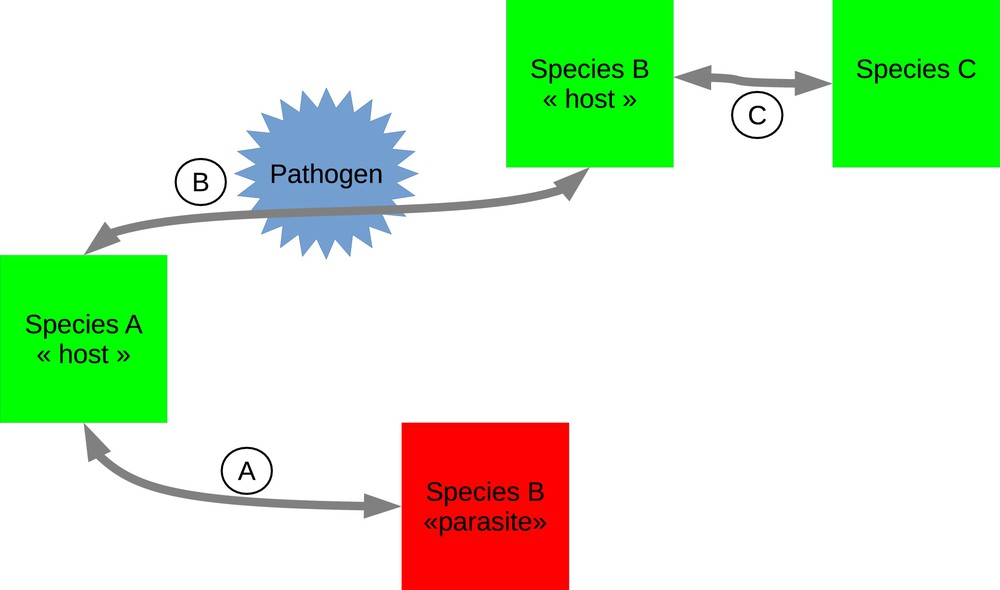
A schematic view of the possible mechanisms for HTs. A. Direct parasitism, like for Striga or Cuscuta. B. Pathogen-mediated transfer allowing the transfer of DNA between host species sharing the same pathogen. C. Grafting.
5 Conclusion
HTs have long been considered as rare phenomena. With the development of new sequencing technologies, HTs can be studied at a large-scale through comparative genomic studies. Several recent studies have evidenced that HTTs are much more frequent than previously thought, which suggests that such gene flows may play a role in the transmission of genetic information among sexually isolated species, even if the exact mechanisms of these transfers are not well understood. The latest sequencing technologies provide new opportunities for ecosystemic genomic studies. These will allow us to unravel the extent and the possible role of HTs in ecosystems.
Disclosure of interest
The author declares that he has no competing interest.


