1 Introduction
Since their origin, hunting and gathering had been the primary mode of subsistence for modern humans. But around 12,000 years ago, humans switched from a hunter-gatherer lifestyle to an agricultural lifestyle. This transition in human behavioural ecology is known as “the Neolithic revolution”. The Neolithic revolution has marked one of the most profound changes in human evolution. With reliable food stocks, human populations have increased, expanded, and built civilizations with environmental and cultural consequences that persist today. One of the primary drivers of this transition is the domestication of plants, a process whereby wild plants have been evolved into crop plants through human-mediated selection. Plant domestication has entailed co-dependency between humans and plants while promoting plant adaptation to a new ecological niche, the field. How complex were domestications? Where did they take place? How long did they last? These are some of the questions at the interface between archaeology, ecology and evolutionary genetics that have been until today actively debated, starting with the observations of Charles Darwin first published in 1868 in a book entitled “The Variation of Animals and Plants under Domestication”.
2 What is plant domestication?
Domestication can be described as a set of consecutive stages that begins with the onset of domestication followed by an increase in the frequency of a set of desirable traits (the domestication traits), and which culminates with the emergence of cultivated populations adapted to both human needs and a cultivated environment. Thereupon a first challenging task is to define a domestication syndrome, which is the subset of traits that collectively form the morphological and physiological differences between crops and their wild progenitors. Domestication traits were the very first targets of early farmers as opposed to traits selected later during crop diversification. We expect them to be fixed or nearly fixed in the cultivated forms as a result of intense human-driven positive selection.
Domesticated traits can be classified into three categories:
- • yield-related traits that affect propagule retention, shape and size – longer and more rigid stolons in cultivated potatoes, loss of seed shattering in cereals, indehiscent pods in legumes, increase in fruit size of cultivated tree species are some examples;
- • food usage-related traits such as reduction of chemical and physical defences, and reduction of propagule ornamentations that facilitate dispersal in the wild – loss of bitterness in cultivated almonds, loss/reduction of awns in rice and wheat fall in this category;
- • cultivation-related traits that concern growth habit and loss of seed dormancy – the determinacy in bean cultivated forms and loss of seed dormancy in chickpea illustrate this last category.
Domesticated plants often rely on human maintenance to ensure their reproductive success, and domesticated traits are usually highly deleterious in the wild environment. For instance, propagule dissemination or seed dormancy are essential for survival in the wild but selected against in the field.
3 Single versus multiple domestications
At least 11 regions of the Old and New World can be considered as independent isolated centres for the origin of crops, several of which occur in Central and South America, Africa, and South East Asia [1]. The Fertile Crescent is considered as the cradle of plant domestication with the emergence of major cereals such as wheat, barley, oats, rye, as well as lentils and chickpeas. Some of the related wild forms of these crops were cultivated before domestication. Hence Weiss et al. [2] have reported consistent evidence of granaries containing hundred thousands of wild barley and oat seeds in the Jordan Valley, suggesting seed management and perhaps mass-selection predating domestication.
While attempting to determine the origins of crops using genetic data, it is not uncommon to arrive at conflicting interpretations. Recurrent gene flow among cultivated forms or between wild and cultivated gene pools, for instance, may mask multiple domestication events. It is therefore important to merge multiple sources of data and assess congruence between archaeological findings and genetic analyses. Paleoclimatic reconstructions may also guide inferences on the ancient niches occupied by wild progenitors as reported for teosinte/maize landraces by Hufford et al. [3]. Along the same line Kraft et al. [4] have integrated evidence from paleobiolinguistics – the presence of words designating the cultivated species in an ancestral language being indicative of its importance – as a geographical grid layer complementary to that of genetic diversity and environmental niche projections in order to help refine the location of chili pepper (Capsicum annuum) domestication in Mexico.
Factors such as the distribution area of the crops wild progenitors as well as the rapidity of crops spread outside their centre of origin have likely contributed to the emergence of the three described alternative domestication scenarios: a domestication event from a single gene pool in a restricted area, the best example so far being maize [5]; a diffuse domestication from wild gene pool(s) distributed in a broader area, pearl millet domestication in the Sahel zone, which illustrates this situation [6], along with barley with the recent discovery that the genome of cultivated barley is a mosaic of several wild source populations [7]; multiple domestications in geographically distinct areas. Examples of the latter include the common bean, which was domesticated independently in Mexico and the Andes from two divergent gene pools [8] as well as Asian rice with two, perhaps even three, independent domestications [9].
4 Determinants of the domestication syndrome
Most domesticated genes so far were detected through the so-called top-down approach from phenotype to genotype. Crosses between wild and cultivated forms and examination of co-segregation of genetic markers and phenotypes in the offspring of these crosses (Quantitative Trait Loci mapping) have recovered a number of candidate regions. Genes in these regions were further identified by a combination of fine mapping, association mapping, and functional analyses including mutant complementation and gene expression assays. Analyses of patterns of polymorphism aiming at seeking footprints of selection in cultivated samples are also often performed to corroborate molecular evidence.
Table 1 presents the current domestication genes/loci list with their corresponding functional annotations. A prime example of a major domesticated gene is the teosinte branched 1 (tb1) gene. First identified from QTL mapping as a major determinant of the differences in inflorescence morphology and plant architecture between maize and teosinte, the construction of a near-isogenic line containing the teosinte QTL in a maize background failed to complement the maize Tb1 mutant allele [10]. The gene cloned via transposon tagging, belonged to the TCP family of transcription regulator. Expression patterns were consistent with the overexpression of the maize allele in the lateral primordia inducing a strong apical dominance with reduced lateral branches and feminization of the lateral terminal inflorescences [11]. Further comparison of the wild and cultivated alleles revealed a drastic reduction of diversity from 5’ UTR to 60–90 kb upstream the gene [12]. More recent work from the same team has revealed selection from standing variation at a Hopscotch transposable element situated in the tb1 regulatory region. This element enhances tb1 expression in the cultivated form.
Selected list of genes/loci whose function/phenotype/selective patterns offer convincing evidence of their involvement in domestication.
| Crop species | Common name | Gene name (abbreviation) | Trait | Gene type | References |
| Brassica oleracea | Broccolia | BoCAULIFLOWER (BoCAL) | Affects floral primordia, alterations in inflorescence morphology | Transcription factor | 1,2 |
| Glycine max | Soybean | SHATTERING1-5 (SHAT1-5) | Increased lignification of fiber cap cells leads to shattering-resistant pods | Transcription factor | 3 |
| Hordeum vulgare | Barley | INTERMEDIUM-C (INT-C) | Fertility of lateral spikelets and tillering | Transcription factor | 4 |
| Hordeum vulgare | Barley | Nud (nud) | Caryopsis with easily separable husks | Transcription factor | 5 |
| Hordeum vulgare | Barley | SIX-ROWED SPIKE (HvVRS1) | Development and fertility of lateral spikelet | Transcription factor | 6 |
| Oryza sativa | Rice | BLACK HULL4 (Bh4) | Changes color of seed hull from black to white | Amino acid transporter protein | 7 |
| Oryza sativa | Rice | GRAIN WIDTH5 (GW5) | Increase of grain size | Polyubiquitin-interacting protein | 8 |
| Oryza sativa | Rice | OsLIGULELESS1 (OsLG1) | Alteration in laminar joint and ligule development forming closed panicles | Transcription factor | 9 |
| Oryza sativa | Rice | OsPROSTRATE GROWTH1 (PROG1) | Tiller angle leads to erect growth (plant architecture) | Transcription factor | 10, 11 |
| Oryza sativa | Rice | Red pericarp (Rc) | Pericarp color | Transcription factor | 10, 12 |
| Oryza sativa | Rice | SHATTERING4-1 (sh4-1) | Reduced seed shattering | Transcription factor | 13 |
| Phaseolus vulgaris | Common bean | PvTERMINAL FLOWER1 (PvTFL1y) | Determinate shoots with a terminal inflorescence | Transcription cofactor | 14 |
| Solanum lycopersicum | Tomato | LOCULE NUMBER (LC) | Increase in the number of locules | Transcription factor | 15 |
| Solanum lycopersicum | Tomato | FASCIATED (fas) | Increase in the number of carpels and locules | Transcription factor | 16 |
| Solanum lycopersicum | Tomato | Fruit weight 2.2 (fw2.2) | Alteration in fruit size | Cell number regulator protein | 17 |
| Sorghum bicolor | Sorghum | SbSHATTERING1 (SbSH1) | Non-shattering of seeds | Transcription factor | 18 |
| Triticum aestivum | Common wheat | Wheat AP2-like (WAP2) (Q) | Allows free-threshing and spelt spike formation | Transcription factor | 19 |
| Zea mays | Maize | BARREN STALK1 (ba1) | Prevents axillary meristem formation | Transcription factor | 20 |
| Zea mays | Maize | Brittle2 (bt2) | Increase in yield and different amylopectin properties | Enzyme | 21 |
| Zea mays | Maize | Grassy tillers1 (gt1) | Suppression of elongation of lateral ear branches | Transcription factor | 22 |
| Zea mays | Maize | PROLAMIN-BOX BINDING FACTOR (PBF1) | Unclear | Transcription factor | 23, 24 |
| Zea mays | Maize | Ramosa1 (ra1) | Branching architecture | Transcription factor | 25 |
| Zea mays | Maize | Starch branching enzyme IIB (ae1) | Amylopectin structure leading to starch pasting properties | Amylose extender | 21 |
| Zea mays | Maize | Teosinte branched 1 (tb1) | Apical dominance, short ear tipped branches | Transcription factor | 26, 27 |
| Zea mays | Maize | Teosinte glume architecture 1 (tga1) | Softer glume leads to kernel exposition | Transcription factor | 28 |
| Zea mays | Maize | Zea agamous-like1 (Zagl1) | Increase in female ear length | Transcription factor | 29, 30 |
| Zea mays | Maize | ZmSHATTERING1 (ZmSh1) | Reduced seed shattering | Transcription factor | 18 |
a Also includes the varieties: Brussels sprouts, Cabbage, Cauliflower, Kale and Kohlrabi.
While Single Nucleotide Polymorphisms are the most frequently reported changes, additional examples present evidence of transposable elements being the causative domestication mutations. Hence, a 4.1-kb retrotransposon insertion in the PvTFL1y gene provokes growth determinacy in common bean [13], and a Helitron insertion is found in barren stalk1 (ba1) maize gene, which regulates together with tb1 vegetative lateral meristem development and patterning of inflorescences [14]. One of the most recent and interesting discoveries of a domestication gene was found by Müller et al., 2016 [15]; it is a 3-bp deletion in the coding sequence of the Arabidopsis EID1 homologous gene in cultivated tomatoes. The domesticated allele noticeably delays the phase of the circadian clock by three hours on average. EID1 controls the network of genes that allows anticipating daily and seasonal changes and better synchronizing physiological processes. The adaptive advantage of the cultivated allele may be linked to the completion of tomato domestication outside its native range where it encountered longer days and evolved light-related damage avoidance [15].
Once causal mutations have been pinpointed, it is inevitable to wonder what kind of genes is most prevalent. Are domestication genes superheroes (structural genes) or masterminds (genes controlling regulatory network readjustments)? So far, most phenotypic changes associated with domestication seem to be orchestrated by mutations in regulatory genes (Table 1). Considering that transcription factors represent ∼ 5% of the genes in the model species Arabidopsis [16], this observation is puzzling and may indicate, as John Doebley [23] pointed out, that domestication is a process of genetic tinkering as opposed to genetic disassembling. In other words, domestication seems to have involved re-orchestration of gene networks and their expression by targeting “masterminds” rather than via the accumulation of null or loss-of-function mutations.
5 Genetic or phenotypic convergence?
Although different species were domesticated in different geographical locations at various times through history, it is possible to identify similar outcomes in their phenotypes which is termed phenotypic convergence – see Tenaillon and Manicacci [17]. It is of interest to pry into the nature of the genetics behind these traits, not only to better understand how adaptation proceeds, but also to address questions about the degree of genetic convergence in the evolutionary paths underlying convergent phenotypes. Was the same set of orthologous genes involved in the acquisition of similar traits among species? Were the same genes targeted by mutations within species when multiple domestications took place?
In most cases, convergence at the genetic level (Fig. 1) has found little support amongst cultivated lineages. Hence, as exemplified with barley non-brittle rachis, several different genes can confer similar phenotypes [18]. Exceptions include recurrent selection of orthologous genes encoding loss of seed shattering at the Sh1 gene in sorghum, and at the OsSh1 and ZmSh1 genes in rice and maize respectively [19]. In the common bean, the genome scan by Schmutz et al. [8] found 59 shared domestication candidate genes between the Mesoamerican and Andean gene pools, representing 3% and 8% of each pool's candidates, respectively. Kwak et al. [20] actually reported independent selection events on the PvTFL1y gene in each common bean gene pool. At a finer scale, different mutations may be observed on the same gene, resulting in similar domestication phenotypes as in the case of rice Bh4 gene that generates white-hulled seeds [21]. Interestingly, such examples of repeated evolution on the same genes or orthologous genes across species are more often observed during crop diversification than during domestication [22].
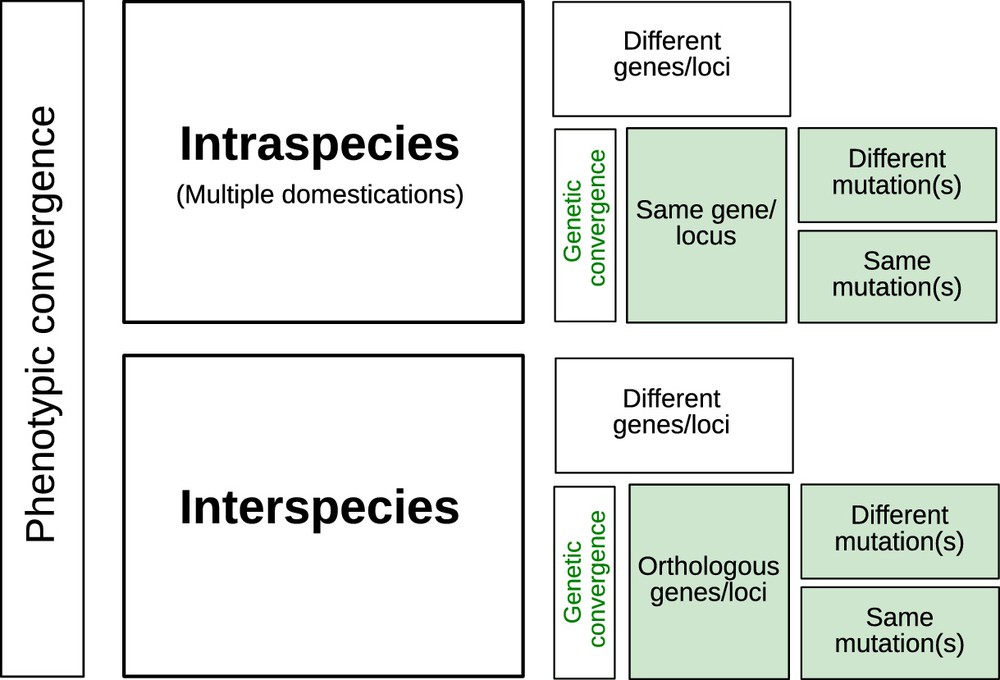
Levels of genetic convergence associated to phenotypic convergence in one or more species.
6 What is the pace of domestication?
First thought to be a rapid process that must have presented an immediate advantage for the early farmers, the domestication process has now endorsed the status of a slow transition from wild to domesticated plants cultivation. Archaeological studies hence report the persistence within a given site of wild and cultivated forms over a long time period with a slow increase of the latter. Hence, Fuller et al. [23] have established that fixation of the non-shattering phenotype in barley, einkorn and emmer extended over a period of 2000 to 2500 years. In rice, there is evidence of a mix of wild (shattering) and cultivated (non-shattering) rice in Chinese sites from the lower Yangtze valley, with a gradual increase in the domesticated forms from 27% (4900 BC) to 39% in 300 years [24]. But such patterns may vary from one site to another: by 6300 BC non-shattering acquisition was already complete in the middle Yangtze, suggesting an accelerated process in this area as compared to the lower Yangtze.
Many factors may influence the pace of domestication across species and sites. For instance, as discussed by Fuller [25], cultural practices related to the harvest of grains have certainly played a major role. Harvesting immature grains in cereals would delay selection for domesticated phenotypes, while storage of late-harvest mature seeds for sowing the following year would instead favour non-shattering phenotypes. Life history traits, in particular annual versus perennial life cycles have also clearly impacted domestication pace. Hence the evolution of perennial cultivated forms is affected by long juvenile periods, high level of gene flow with wild relatives, and somatic mutations transmitted by clonal propagation [26]. The rate of adaptation to the cultivated environment is also dictated by the mating system, which influences the fixation time of beneficial mutations. Glémin and Ronfort [27] have demonstrated that this rate is shorter in selfers than in outcrossers when adaptation proceeds through recessive or partially recessive mutations; a recessivity expected for domesticated traits that are most likely highly deleterious in the wild. Note that the deleterious effect of such alleles must also contribute to maintain them at very low frequency, which makes the selection from standing variation less likely. Selfing is also an efficient way to protect domesticated forms from recurrent maladaptive gene flow from sympatric wild forms. Finally, population size interferes with the aforementioned predictions by modulating the efficacy of selection. Overall, domestication must have proceeded faster in selfers than outcrossers and faster in large population size. The first prediction is consistent with a majority of selfers found among domesticated crops [27].
7 Consequences of domestication for the genetic diversity crop
The most notable consequence of domestication is a loss of genetic diversity. This has been observed in many species and varies from few percents to 20% up to 80% loss at the nucleotide level in maize [28] and durum wheat respectively [29]. Domestication is a recent enough process to detect the footprints of what is commonly called, the domestication bottleneck, a direct consequence of selection on a subset of wild individuals/populations. This bottleneck is likely underestimated because of the recovery of diversity since domestication through mutations, population expansion, and gene flow from wild relatives. Bottleneck scenarios have been modelled in multiple domesticated species, but the impact of gene flow has been overlooked. While there is evidence of recurrent gene flow between wild and domesticated forms, a compilation suggests that the majority of crops actually possess reproductive barriers, 38% of them being linked to either ploidy differences or reduced hybrid fitness [30]. Whether these barriers can be considered as a domestication trait is still an open question.
Both shrinks in population size and to a lesser extent impact of selective sweeps on neighbouring pre-existing variations [31] have inflated the accumulation of slightly deleterious mutations, an effect magnified in poor recombining regions [32]. There is hence a cost to domestication. It can be estimated by analysing the enrichment of nonsynonymous to synonymous derived substitutions in the cultivated form with respect to the wild form. Nabholz et al. [33] have found good evidence for such enrichment in the African rice (Oryza glaberrima) in comparison to its wild progenitor Oryza barthii and further showed that it is more pronounced in regions suffering strong drift.
8 Conclusion
Domestication studies continue to be a fascinating ground to delve into. By combining approaches from diverse disciplines, the origins and processes accompanying crop domestication have begun to be understood. So far, research on the genetic unravelling of domestication points to modulation in the expression of mastermind genes, which in turn exert a downstream rewiring of genetic networks. Hitherto, convergence at the gene level among crops or between crops independent domestications has rarely been observed. In fact, because the pace of domestication is influenced by many intricate factors related to life history traits, population size and trait genetic determinism in combination with cultural practices, the emerging domestication patterns are truly species-specific. They span very slow to rapid transitions embedded in single or multiple domestication events. Conversely, a consequence of domestication that has been recurrently encountered is a loss of genetic diversity that stresses the importance of assessing the functional variation of wild genetic resources to broaden the usable genetic diversity in conventional breeding programs.
Disclosure of interest
The authors declare that they have no competing interest.
Acknowledgements
We thank Georges Pelletier, Bernard Dujon and André Gallais for useful comments on the manuscript as well as all the superheroes and masterminds cited in this review for their insightful and inspiring work. The work of M.I.T is supported by the “Agence nationale de la recherche” (Project ANR 12-ADAP-0002-01) and N.E.M-A is funded by a CONACYT scholarship (579966/410748).


