1 Introduction
There is growing evidence that crude oil is degraded in situ by anaerobic prokaryotes. Heavy oils, tar and gas are the legacy of this microbial degradation over geological times. Petroleum reservoirs are important habitats within the deep biosphere, in which microbial consortia live at the oil–water interface, despite the drastic physicochemical conditions in the reservoirs, e.g., temperatures up to 190 °C in the Elgin–Franklin fields, North Sea [1], salinities up to 400 g/l in the Verkhnechona field, Russia [2], or the lack of oxygen. Many anaerobic prokaryotes have been retrieved both by molecular and cultural approaches from such ecosystems [3]. The microbial mineralization of OM relies on the cooperation of different groups of anaerobic microorganisms in sequence: fermenters, syntrophs, and methanogens [3]. This microflora is composed of genera belonging to the bacterial phyla Firmicutes, Bacteroidetes, and Proteobacteria and methanogenic archaea, such as Methanomicrobiales, Methanosarcinales, and Methanobacteriales [4]. The bacterial diversity in oil reservoirs often presents large populations of Epsilonproteobacteria [5,6]. Epsilonproteobacteria are major sulfur-oxidizers, nitrate reducers autotrophs and mixotrophs, whose contribution to the sulfur cycle is essential in different environments, such as deep-sea hydrothermal vents and sediments [7], sulfidic cave springs [8], or episymbiotic associations [9]. In these environments, the diversity of Epsilonproteobacteria is often dominated by a single genus: Sulfurovorum in sulfidic biofilms, Sulfurimonas in the Colleville oil field, Arcobacter in the Pelican Lake oil field or Sulfuricurvum in the Athabasca oil sand reservoir. Although Epsilonproteobacteria are frequent, or dominate the bacterial diversity in different oil environments, it is unlikely that they partake in the breakdown of the organic matter of the oil, but more likely oxidize the fraction of sulfur compounds of the oil [5,6].
In the absence of the appropriate geologic structure to form a reservoir, crude oil and gas formed in the depth naturally seep out at the surface. These oils are often heavily degraded as a result of microbial activity before or during the migration to the surface, which raises questions about whether the oil-degrading prokaryotes migrate to the surface along with the oil, and whether petroleum degradation persists at the surface. To address this question, we investigated the microbial populations at the Puy-de-la-Poix, a small, naturally flowing seep of heavily degraded oil, or bitumen, in the Limagne region (France). We show here that as observed in other degraded-oil environments, the bacterial diversity at the Puy-de-la-Poix was characterized by a large majority of Epsilonproteobacteria of the Sulfurimonas genus.
2 Methods
Sampling: The “Puy-de-la-Poix” is a naturally flowing tar source located near the airport of Clermont-Ferrand (France, 45.7822 N 3.14642 E, 340 m, Fig. 1). The source appears as a small pool of ca. 3 m by 2 m, which may be covered by rain fall water according to the season, and overflows into the adjacent agricultural fields. At the time of sampling, the air temperature was 6 °C. The tar formed a thick mass on top of the source and the water covering the tar was ca. 2–5 cm deep. The water above the tar was sampled aseptically using sterile syringes (100 ml, surface water samples, SW). The remaining surface water was removed to avoid cross contamination with waters below the tar. Underground waters (UW, 100 ml) were sampled below the tar using sterile tubing punched through the tar. The chemical composition of the underground water can be found in Table S1. A 3-cm-thick, 25 × 25 cm square piece of bitumen was cut aseptically with a sterile blade (bitumen sample, B).
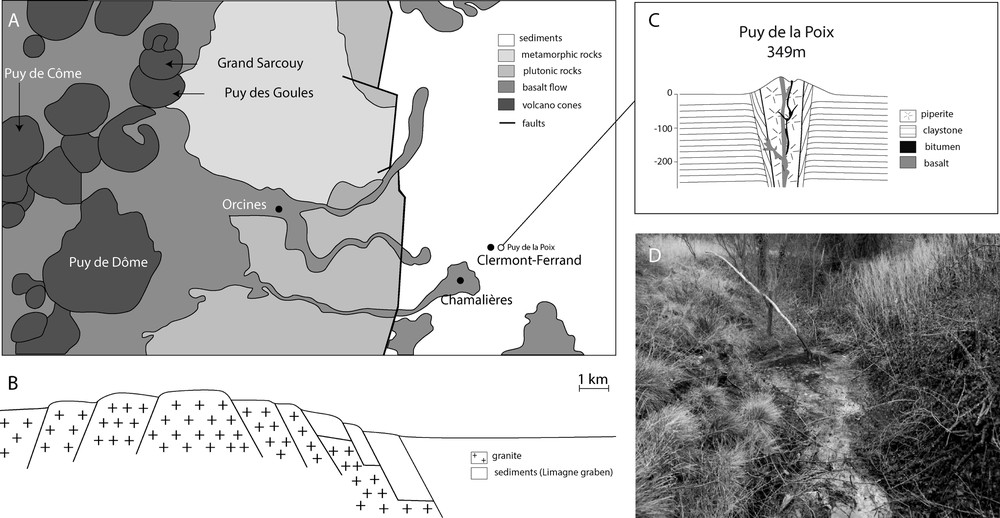
Geographical and geological characteristics of the Puy-de-la-Poix. Panel A: Geologic map close up of the Clermont-Ferrand area. The Puy-de-la-Poix is located east of the city, next to the airport. Panel B: Corresponding cross section. Panel C: detailed geologic structure at the Puy-de-la-Poix Panel D: The seep flows from the top of the ancient volcano into the nearby agricultural fields.
16S library construction: Metagenomic DNA was prepared from 1 g of bitumen or 50 ml of water sample as described previously [10]. The complete 16S rRNA gene was amplified using the universal eubacterial primers pA and pH [11] and cloned into pGEM-T easy (Stratagene, France) to generate the UW, B, and SW libraries. The complete sequence of the 16S gene was determined by Sanger sequencing. GenBank accession numbers for representative groups of Epsilonproteobacteria are JX403021-JX403066.
Diversity analyses: Sequences were classified using the RDP classifier with a confidence threshold of 80% (https://www.rdp.cme.msu.edu). Mothur 1.36 [12] was used to eliminate chimera (chimera.uchime), assign sequences to OTUs (align.seqs, dist.seqs, cluster, OTUs, 97%), get representative sequences (get.oturep), and calculate diversity values (HShannon and SChao1). Phylogenetic trees were reconstructed with Seaview using PhyML, and supported by 1000 boostrap repetitions [13].
Cultivation: Growth was assayed in media specific for Epsilonproteobacteria: Thiomicrospira medium DSM142, Sulfurospirillum medium DSM771 or Sulfurimonas medium DSM1053 (http://www.dmsz.de). Growth media were supplemented with electron donors (lactate, acetate, fumarate, formate, CO2) and acceptors (SO2, SO4, NO3, SeO3, thiosulfate) at 5 mM final concentration. Twenty-milliliter enrichment cultures were set in sealed 50-ml serum vials under nitrogen atmosphere. Enrichment cultures were started with a 200-mg piece of bitumen. Cell counts were determined at least twice a week using a Thoma cell. Positive cultures were subcultured by serial dilution every two weeks until pure cultivable isolates were obtained. Purity was estimated by visual inspection of the culture and PCR amplification, cloning and sequencing of the 16S rRNA gene.
3 Results and discussion
Bacterial diversity at the Puy-de-la-Poix. A total of 159 16S rDNA bacterial sequences, e.g., 58, 49 and 52 clones from the bitumen (B), underground water (UW) and surface water (SW) libraries, respectively, were obtained after quality and chimera screening. The sequences clustered into 48 different OTUs, belonging to six phyla (Table S2). The diversity is dominated by proteobacteria, e.g., 96,5%, 93,9%, and 84,6% in the B, UW, and SW libraries, respectively. The community diversity and richness is lowest in the bitumen (Table 1). Community richness is similar in both water bodies, although the Hshannon index evidences a higher diversity in the surface water. The higher diversity is associated with drastically different bacterial communities. Sequences of the SW library are composed of equal fractions of alpha-, beta-, epsilon and gammaproteobacteria (Fig. 2), while in the UW and B libraries are dominated by Epsilonproteobacteria (80.4 and 83.9% respectively). Most Epsilonproteobacteria sequences at Puy-de-la-Poix are associated with the genus Sulfurimonas, which represent 56.9% and 63.3% of the total bacterial diversity in the B and UW libraries, respectively. The remaining diversity of Epsilonproteobacteria is represented by Sulfurovum and Arcobacter (12.1% and 10.3% of the total bacterial diversity respectively in the B library, and 8.1% of the total bacterial diversity for both groups in the UW libraries). A single Sulfurimonas OTU, e.g. OTU 11 (Table 2, Fig. 2) dominates Epsilonproteobacteria diversity and accounts for 30 and 35% of Epsilonproteobacteria in the UW and B libraries (Fig. 2), but is not found in the SW library. OTU 11 is most closely related to the type species of the Sulfurimonas gotlandica species [14]. Four OTUs were associated with the Sulfurovum aggregans type strain Monchim33, which has been isolated from a hydrothermal vent system [15]. The Sulfurovum OTUs are related to environmental clones of very diverse origins, a soil, a sediment mesocosm, a bivalve symbiont or an acid cave. Arcobacter sequences (6 OTUs) represent a minor, but significant, component of the bacterial diversity (10% and 8% for the B and UW libraries respectively). The last two OTUs belong to the Sulfuricurvum and Sulfurospirillum genera (Table 2). There is a large overlap between the bacterial populations of the UW and B libraries, which is consistent with a common origin of the two populations, and the probability that Epsilonproteobacteria were present in the zone of contact of the oil and water. Indeed, such large prevalence of Epsilonproteobacteria has been reported previously at the oil/water interface in other geological settings such as a highly degraded oil reservoir or an oil spill from a storage tank [5,6]. The absence of a bacterial diversity related to known bacterial fermenters or syntrophs at the Puy-de-la-Poix, and the low level of sulfate-reducers suggested that the degradation of the organic matter of the oil and sulfate reduction may not be occurring at a significant level at the Puy-de-la-Poix, and that microbial degradation occurred during the transit of the oil to the surface.
Community richness (Schao1) and community diversity (HShannon).
| S Chao1 | H Shannon | |
| B | 60 | 2,57 |
| UW | 306 | 2,78 |
| SW | 335 | 3,35 |
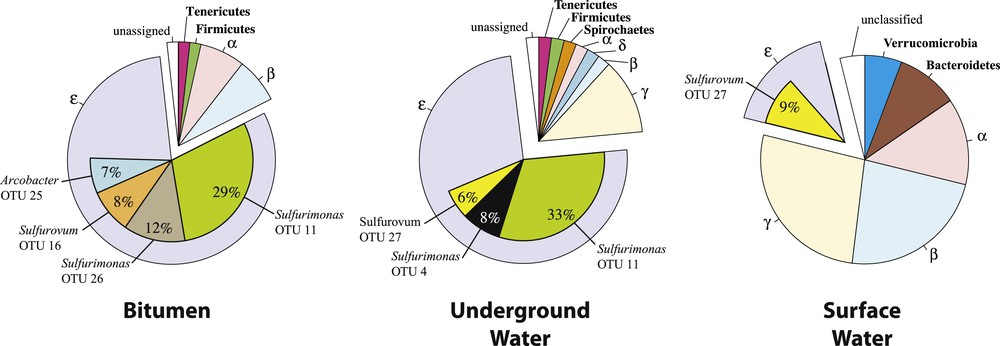
Total bacterial diversity at the Puy-de-la-Poix. Major Epsilonproteobacteria OTUs and their taxonomy, are shown as inserts within the Epsilonproteobacteria (purple).
Diversity and taxonomic position of the epsilonproteobacterial sequences.
| OTU | Reference clone | B | UW | SW | Identification | Closest cultivated isolatea | Closest environmental sequenceb | ||||
| Species and strain name | Sampling site | % ID | Accession | % ID | Sampling site | ||||||
| 1 | UW-31 | 1 | Arcobacter | Arcobacter cloacae SW28-13 | Sewage station | 98.9 | |||||
| 8 | B-31 | 1 | Arcobacter | Arcobacter marinus CL-S1 | Seawater | 99.4 | AY569293 | 99.9 | Hot spring | ||
| 9 | UW-55 | 1 | Arcobacter | Arcobacter bivalviorum F4 | Mussels | 92.7 | FR666865 | 97.3 | Cold seep | ||
| 12 | UW-35 | 1 | Arcobacter | Arcobacter venerupis F67-11 | Mussels | 95.8 | HQ538628 | 98.1 | Sludge | ||
| 14 | B-2 | 1 | Arcobacter | Arcobacter nitrofigilis DSM 7299 | Marshplant roots | 98.6 | AY704399 | 99.6 | Crustal fluids | ||
| 25 | B-9 | 4 | Arcobacter | Arcobacter venerupis F67-11 | Mussels | 97.1 | EU617863 | 98.3 | Yellow Sea sediment | ||
| 2 | UW-36 | 1 | Arcobacter | Arcobacter ellisii F79-6 | Mussels | 92.7 | EU265974 | 97.2 | Lake sediment | ||
| 10 | UW-58 | 1 | Sulfuricurvum | Sulfuricurvum kujiense DSM 16994 | Oil polluted water | 98.7 | |||||
| 6 | UW-89 | 1 | Sulfurimonas | Sulfurimonas sp. MA01 | Marine sediment | 97.0 | AB240698 | 97.8 | Sediments | ||
| 4 | UW-45 | 1 | 4 | Sulfurimonas | Sulfurimonas autotrophica DSM 16294 | Marine sediment | 95.1 | AY922199 | 98.6 | Various | |
| 7 | UW-33 | 1 | Sulfurimonas | Sulfurimonas autotrophica DSM 16294 | Marine sediment | 93.6 | EF219001 | 95.9 | Hydrothermal vent | ||
| 13 | B-18 | 1 | Sulfurimonas | Sulfurimonas denitrificans DSM 1251 | Hydrothermal vent | 91.2 | EU570862 | 93.3 | Lake sediment | ||
| 24 | B-32 | 3 | 1 | Sulfurimonas | Sulfurimonas denitrificans DSM 1251 | Hydrothermal vent | 95.1 | FJ717065 | 97.5 | Mesocosm | |
| 29 | B-1 | 1 | Sulfurimonas | Sulfurimonas denitrificans DSM 1251 | Hydrothermal vent | 96.1 | UOU46506 | 98.2 | Oil field | ||
| 22 | B-54 | 1 | 1 | Sulfurimonas | Sulfurimonas denitrificans DSM 1251 | Hydrothermal vent | 94.0 | DQ112511 | 95.8 | Mudflat sediment | |
| 3 | UW-88 | 1 | Sulfurimonas | Sulfurimonas gotlandica GD1 | Pelagic redoxcline | 95.1 | AB478650 | 97.7 | Microbial mat | ||
| 5 | UW-41 | 1 | Sulfurimonas | Sulfurimonas gotlandica GD1 | Pelagic redoxcline | 85.6 | FJ628185 | 89.1 | Lake sediment | ||
| 11 | B-22 | 17 | 16 | Sulfurimonas | Sulfurimonas gotlandica GD1 | Pelagic redoxcline | 94.8 | AB478672 | 96.6 | Microbial mat | |
| 18 | UW-84 | 1 | Sulfurimonas | Sulfurimonas gotlandica GD1 | Pelagic redoxcline | 93.5 | FJ037617 | 97.0 | Iron-ixodizing biofilm | ||
| 19 | B-7 | 2 | 1 | Sulfurimonas | Sulfurimonas gotlandica GD1 | Pelagic redoxcline | 94.6 | FJ437845 | 98.0 | Lake sediment | |
| 20 | SW-37 | 1 | Sulfurimonas | Sulfurimonas gotlandica GD1 | Pelagic redoxcline | 94.8 | FJ437845 | 97.7 | Lake sediment | ||
| 21 | UW-52 | 1 | Sulfurimonas | Sulfurimonas gotlandica GD1 | Pelagic redoxcline | 94.5 | AB478672 | 97.7 | Microbial mat | ||
| 26 | B-16 | 7 | Sulfurimonas | Sulfurimonas paralvinellae GO25 | Polychaete nest | 95.1 | AB247901 | 96.4 | Hydrothermal vent | ||
| 28 | UW-29 | 1 | Sulfurimonas | Sulfurimonas gotlandica GD1 | Pelagic redoxcline | 91.3 | GQ261791 | 94.0 | Deep sea | ||
| 23 | SW-32 | 1 | Sulfurospirillum | Sulfurospirillum arsenophilum MIT-13 | 98.6 | ||||||
| 16 | B-13 | 5 | Sulfurovum | Sulfurovum aggregans Monchim33 | Hydrothermal vent | 95.9 | GU583971 | 97.6 | Mangrove Soil | ||
| 17 | B-12 | 1 | 1 | 1 | Sulfurovum | Sulfurovum aggregans Monchim33 | Hydrothermal vent | 96.8 | FJ717160 | 98.6 | Mesocosm |
| 27 | UW-37 | 3 | 5 | Sulfurovum | Sulfurovum aggregans Monchim33 | Hydrothermal vent | 95.3 | EF467592 | 100 | Acid cave | |
| 15 | B-26 | 1 | Sulfurovum | Sulfurovum aggregans Monchim33 | Marine sediment | 94.1 | EU487914 | 95.9 | Bivalve-symbiont | ||
| L12 | Sulfurospirillum | Sulfurospirillum multivorans DSM 12446 | Activated sludge | 93.5 |
a Taxonomy, strain name and percent homology of the 16S rRNA gene sequence from the closest cultivated organisms.
b Accesssion number and percent identity of the closest 16S rRNA gene sequence from uncultured organisms. This information is only provided when sequence more closely related than the cultivated isolate.
Isolation of cultivableEpsilonproteobacteria. Obtaining cultivable Epsilonproteobacteria from the Puy-de-la-Poix was a prerequisite to assay their metabolic abilities. Six pure cultures were obtained using the Thiomicrospira mineral base medium supplemented with either acetate or lactate, and nitrate as an electron acceptor. The 16S rDNA gene sequences of the six isolates were almost identical (> 99% sequence identity), defining a single OTU closely related to Sulfurospirillum multivorans (Clone L12, Table 2 and Fig. 3). The strains isolated here are closely related, but do not belong to OTU 23 (SW library, Table 2, Fig. 3). Microscopic observations of the isolates revealed curved or helical rods. The cells were non-motile. Growth was obtained with acetate and lactate, but not pyruvate, formate or sugars in the presence of nitrate under anaerobic conditions. No growth was observed on sulfate or thiosulfate as electron acceptors. As expected, no growth occurred under aerobic conditions. Typical growth was low, reaching ca. 107 cells/ml within two weeks. The apparent doubling time of strain L12 is ca. 24 h. This doubling time is extremely long for the Sulfurospirillum species, for which the doubling time is usually in the order of a few hours. Growth yields were also extremely low. Both observations indicate that the growth medium used to isolate strain L12, although supporting growth, is lacking some important components, which we have been unable to identify thus far. No pure culture belonging to the major Epsilonproteobacteria clades (Fig. 3) could be isolated.
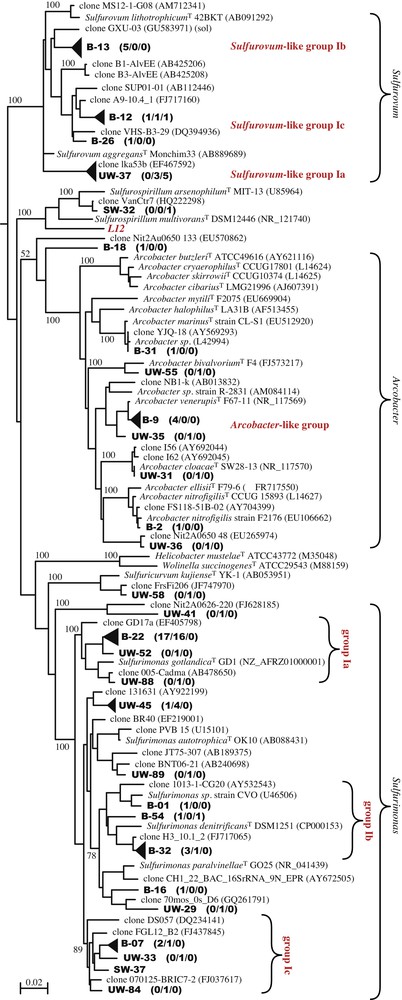
Phylogenetic tree of the 16S rDNA gene sequences from environmental clones of the UW, SW and B libraries and reference sequences from the databases. Clones from the present study are shown in boldface type. The topology was obtained by comparing nearly full-length sequences of the 16S rRNA gene using the maximum likelihood approach with PhyML. Similar tree topologies were produced using other tree reconstruction algorithms. The scale bar represents the number of changes per nucleotide position. Filled triangles were substituted to individual clones for some clusters of sequences to improve readability of the tree. The height of the triangles is drawn to scale with the diversity of each group.
Role ofEpsilonproteobacteriaat Puy-de-la-Poix. Only two OTUs observed at the Puy-de-la-Poix are related to isolates from oil/water interfaces: OTU 10 is related to Sulfuricurvum kujiense, a species isolated from an oil storage tank [16]. The second OTU is related to the CVO strain, which has been isolated from the Coleville oil field [17]. Both S. kujiense and CVO are able to reduce nitrate with the concomitant oxidation of sulfur compounds. Both organisms have been found to dominate the bacterial diversity at their isolation sites, although they were unable to degrade the organic matter of the oil. Therefore, their role in these particular ecosystems, and the source of carbon supporting their growth remained unclear. Most of the Epsilonproteobacteria at Puy-de-la-Poix were most closely related to Sulfurimonas gotlandica [14]. S. gotlandica is a sulfur oxidizer, nitrate-reducing chemolithoautotrophic bacterium that has been isolated from the deep-sea abyssal sediments of the Gotland depth. Similarly to S. kujiense and strain CVO, S. gotlandica is unable to use small organics, such as pyruvate, lactate of formate, but uses dissolved carbonates to support its growth, a phenomenon known as high dark CO2 fixation [18]. Thus, it is very unlikely that Epsilonproteobacteria at Puy-de-la-Poix may be involved in the breakdown of the complex organic matter of the tar. High dark CO2 fixation is important in the suboxic to sulfidic transition zones of aquatic pelagic redox zones, as in the Black Sea and the Baltic Sea from which S. gotlandica has been isolated [14,18–21]. There are enough carbonates in the water at Puy-de-la-Poix to support high dark CO2 fixation by chemolithoautotrophic Epsilonproteobacteria (Table S2) oxidizing sulfur and reducing nitrates. Thus, the dominance of Epsilonproteobacteria could be explained by the refractory nature of the residual organic matter composing the tar, which would limit the access of fermenters and syntrophs to carbon and energy sources, while Epsilonproteobacteria could rely on reduced sulfur compounds and CO2 fixation for growth. Epsilonproteobacteria could reduce sulfur compounds of the oil, but more probably dissolved reduced sulfur species, such as hydrogen sulfide, which are present in the water due to the hydrothermal activity of this geologic province.
4 Conclusions
The bacterial diversity at Puy-de-la-Poix shows an unexpected domination of Epsilonproteobacteria. The lack of known sulfate-reducers or syntrophs is a clear indication that oil mineralization to methane does not occur at this site. The abundance of Epsilonproteobacteria indicates that reduced sulfur species are the major energy source, ruling out significant degradation of the oil organic matter under anaerobic conditions. Our observations do not rule out the possibility of aerobic microbial oil degradation, in contact with surface waters, or when oil overflows in the nearby fields. The presence of large populations of Epsilonproteobacteria at the water/oil interface of different geological settings (oil reservoirs, oil contaminated waters or natural seep) is a clear indication that this interface constitutes a favorable ecological niche for these organisms.
Acknowledgments
The authors would like to thank Pierre Thomas (ENS Lyon) for introducing us to the oil seeps of the Limagne basin. This work was supported in part by a grant from the “École normale supérieure de Lyon” and in part by the “Agence nationale de la recherché” (ANR-10-BLAN-1752-01 Living deep).


