1 Introduction
Ericaceae are considered the eighth largest family of flowering plants, with more than 125 genera and about 4100 species distributed throughout the world [1–3]. In Morocco, the Ericaceae family is represented by only three genera and 10 species including Arbutus unedo, Calluna vulgaris, and Erica spp. [4,5]. The Ericaceae are generally distributed on non-calcareous soils in forests, scrubland and desert regions as well as in the high mountains. With the exception of Arbutus unedo L., which has a wide distribution across Morocco from the western Anti Atlas to the Rif, the other species are quite common only in northern Morocco and sub-humid regions of Tangier and Rif [4,6]. These habitats have soils with low levels of mineral nutrients, acidic pH, poor or free drainage, and are usually climatically diverse [7]. The characteristic harsh edaphic conditions are regarded as the best ecological habitat for most ericaceous shrubs [8].
Ericaceous shrubs can establish root–fungus associations with several fungal partners belonging to different taxa [9–12]. Such multiple symbiotic interactions occur mainly with Ericoid mycorrhizal (ErM) fungi [13–19] and ectomycorrhizal (EcM) fungi. However these interactions are still under debate, actually very little is known about these symbiotic associations. Some authors have reported the ability of ectomycorrhizal (EcM) fungi to form ericoid mycorrhizae [18,20–22], while others suggest that EcM basidiomycetes detected in Ericaceae roots do not form functional ericoid mycorrhizae [23–26].
In addition to ErM fungi, ericaceous plants in both the northern and southern hemispheres can form associations with the most studied group of fungal root endophytes belonging to the group of Dark Septate Endophytes (DSE) [27–29]. Common associations have been reported especially with the group of Phialocephala fortinii s.l.–Acephala applanata species complex (PAC) [30–33]. The mycorrhizal status of this group is still under evaluation; some studies reported that it has neutral or positive effects on plant growth [34–38], while others reported negative results [33,39]. Significantly, some DSE seem to form structures resembling ericoid mycorrhizae in ericaceous roots; however, they have negative effects on functional aspects and plant growth response to colonization [33]. Furthermore, ericaceous plants can also be colonized by fungal pathogens, or saprophytes [40].
Fungal diversity in plant roots is determined by specificity or preference for plant–fungi associations [41,42]. Fungal species with high host preferences, such as mycorrhizal fungi and some endophytic fungi, are expected to be highly influenced by host genetics [40]. This hypothesis is still under evaluation. Studies on the host preference for ErM fungi are few and most of them have suggested the absence of host preference [11,43]; in contrast other authors have indicated the importance of the host in structuring ectomycorrizal communities [44], arbuscular mycorrhizal communities [45,46] and ErM communities [20,40,47].
The diversity and abundance of some species of Ericaceae in Morocco, especially Erica spp., the diversity of the fungi that colonize the root systems of Ericaceae and the importance of these associations in the life cycle of these shrubs, require a more complete characterization of these fungal communities, especially where there is lack of similar studies.
The primary goal of the study was to characterize fungal communities associated with a range of indigenous ericaceous species of Morocco including C. vulgaris, Erica arborea, Erica australis, Erica umbellata, Erica scoparia, Erica multiflora, and A. unedo. To complete the study, results were compared to ericaceous shrubs indigenous to a contrasting ecosystem – the Massif Central in France, with Vaccinium myrtillus and C. vulgaris. The secondary goal was to examine whether or not the fungal communities associated with ericaceous roots diverged among hosts from each region and to show the possible association linking the host species to fungal communities.
2 Material and methods
2.1 Study sites
Ericaceous plants were collected from four contrasting sites in Morocco. The selection of different sites was supported by the presence of different species of ericaceous plants. The maximum of different ericaceous species was found in the sites located in the North of Morocco: Bab berred (B), Mellousa (M), Sahel (S). One site; Ourika (O) located in the south of Morocco was only represented by A. unedo, whereas one site, Loge (L), in the Massif Central in France, was represented by V. myrtillus and C. vulgaris. The study sites were characterized by distinguishable climates (Table 1).
Properties of the plots sampled for ericaceous species sites in Morocco and France.
| Site 1 | Site 2 | Site 3 | Site 4 | Site 5 | |
| Latitude | 35°00′11.65″N | 31°23′36.53″N | 35°45′56.66″N | 35°17′12.79″N | 46°00′19.50″N |
| Longitude | 4°53′51.38″W | 7°45′31.92″W | 5°35′39.79″W | 6°02′55.48″W | 3°47′09.06″E |
| Regions | Bab Berred | Ourika | Mellousa | Sahel | Loge |
| Ericaceous species |
Arbutus unedo
E. arborea |
Arbutus unedo |
E. arborea
E. australis E. multiflora E. umbellata C. vulgaris |
E. scoparia
V. corymbosum |
C. vulgaris
V. myrtillus |
| Elevation (m ASL)a | 1388 | 1115 | 377 | 188 | 1082 |
| Climate | Humid | Semi-arid; sub-humid | Sub-humid | Humid; sub-humid | Mountain climate |
| Pluviometry (mm) | 850 | 450–650 | 700 | 775 | 800 |
| Phosphorus (ppm) | 0.27 | 4.19 | 0.7 | 3.24 | 5.83 |
| Total nitrogen (ppm) | 0.071 | 0.014 | 0.048 | 0.075 | 0.078 |
| pH (H2O) | 5.05 | 7.13 | 4.96 | 5 | 5.3 |
a Above sea level.
The botanical classification of different sampled plants was carried out at the National Herbarium in Rabat. Roots from three plants of each species were sampled in all sites and soil samples from each plot were randomly collected and analyzed for pH, total nitrogen (N) [48], total phosphorus (P) [49] (Table 1).
2.2 Sampling and isolation of fungal strains
The roots of all ericaceous species except the A. unedo were carefully washed with deionized water and surface sterilized separately. The roots went through sequential surface sterilization in diluted (70%) absolute ethanol (0.5 min), sodium hypochlorite (1.65%) (0.5 min), and were then rinsed in sterile deionized water (5 min). The roots of A. unedo were surface-sterilized in diluted (70%) absolute ethanol (5 min), sodium hypochlorite (1.65%) (NaClO) (0.5 min), absolute ethanol (0.5 min) and rinsed in sterile deionized water (5 min).
Three to five surface-sterilized root pieces (1 cm) were then placed onto modified Melin Norkrans Agar media (MMN) [50] in a 9-cm-diameter Petri dish (Fig. 1) and incubated at 25 °C in the dark. Mycelia growing out of the roots were transferred. Cultures were checked for sporulation and slow growing. Sporulating fungi mainly belonging to the anamorphic genera determined by vegetative characteristics and non-sporulating representative isolates were divided into six different morphological groups. Cultures were roughly grouped based on color, appearance and growth rate on potato dextrose agar (PDA), malt agar (MA), and modified Melin–Norkrans agar (MMN). Eighty-four (84) isolates from different morphological groups and habitats were used for molecular analyses.
Surface-sterilized root pieces in a Petri dish.
2.3 Molecular determination of fungi (DNA extraction and ITS amplification)
Fungal DNA was extracted from 50 to 150 mg fresh mycelia using wizard Genomic DNA Purification Kit® (Promega). Amplification of the ITS rDNA regions was performed using the primer pairs ITS1 and ITS4 [51] and the GoTaq® DNA Polymerase kit (Promega) following the manufacturer's instructions. The PCR cycling parameters used were an initial denaturation step for 3 min at 94 °C, then 35 cycles with denaturation at 94 °C for 30 s, annealing at 55 °C for 30 s and extension at 72 °C for 45 s, with a final extension at 72 °C for 10 min. PCR products were checked for length, quality and quantity by gel electrophoresis (1.5% agarose in 0.5% TAE) and double direction sequenced by Eurofins MWG GmbH (Ebersberg, Germany), using the same primers pair. All sequences were corrected and assembled using Chromaslite v2.1.1 (Technelysium Pty). Multiple alignments were first performed with MUSCLE on Phylogeny.fr [52] before using MEGA 4 [53] for NJ analyses with the composite likelihood method.
The resultant ITS sequences were subjected to BLAST searches (GenBank, NCBI) to retrieve the most similar hits. Most ITS sequences had a match above 99% sequence identity and could be assigned to particular species. The rDNA ITS sequences determined in this study have been deposited in the GenBank database under accessions No. KU986751–KU986834.
2.4 Statistical data analyses
Five regions were considered in this study and the number of ericaceous species differed from one region to another (total of 12 species in all regions). Three plants were sampled for each ericaceous species, hence the total number of samples (plants) was 3 × 12 = 36. The presence/absence of fungi belonging to nine fungal orders (eight known and one unknown) was observed on the 36 samples.
The combinations of the three variables: regions, fungal orders and ericaceous species were used to check the hypothesis of the existence of a possible association between any pair of the three variables using the Pearson chi-square test of independence. The magnitude of the association between the three qualitative variables was measured by Cramer coefficient, having 23 categories: five regions, nine ericaceous shrubs, and nine fungal orders were assessed with multiple correspondence analysis (MCA). All the statistical analyses were carried out using the SAS/STAT 9.1 Package [54]. The frequency distribution of the fungi in the different host individuals or regions is given in Tables A.2–A.5 (supplementary material).
3 Results
3.1 Identification of ericaceous species
Six species of ericaceous plants have been identified in the National Herbarium (Rabat, Morocco): C. vulgaris (RAB 78192), E. arborea (RAB 78194), E. australis (RAB 78190), E. umbellata (RAB 78193), E. scoparia (RAB 78229), and E. multiflora (RAB78228). The study sites were located at different altitudes; they have relatively the same climate and pluviometry, except site 2. The soil characteristics differed significantly among sites as well and showed contrasting situations. The pH values were low, attesting to acidic soils (except site 2 where the pH was neutral), most sites displayed low nitrogen and phosphorus levels (Table 1).
3.2 Characteristics of fungal endophytes
Seven hundred and eighty-seven (787) fungal isolates were obtained from roots segments (100 segments per plant) of ericaceae species. Isolates were classified into two groups: one with sporulating fungi mainly belonging to the anamorphic genera determined by vegetative characteristics (Fusarium spp., Penicillium spp., Cladosporium spp. and Alternaria spp.) according to Botton et al. [55], one with non-sporulating isolates which were predominantly dark-colored, ranging from gray to black olive, and from light to dark brown with hyphae showing simple septa, and sterile mycelia. Table A.1 (Supplementary material) and Fig. 2 give examples of the morphological and cultural features, and the growth rate of six distinct non-sporulating fungi.
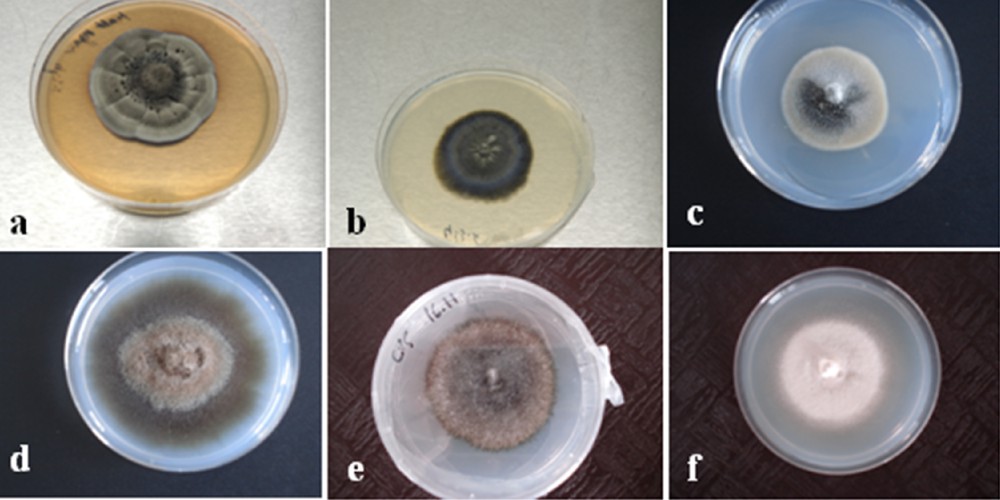
Slow-growing isolates from Ericaceae members grown on a modified Melin–Norkrans (MMN) medium. a: ER2M, b: ER28M, c: ER50M, d: ER37M, e: ER53M, f: ER55M.
Morphotype 1 consisted of cultures with smokey gray to black colonies with white margins; morphotype 2 contained gray to green olive isolates, morphotype 3 contained isolates with smokey gray colonies, morphotype 4 consisted of cultures with light brown to dark brown, while morphotype 5 contained olive black isolates and morphotype 6 consisted of cultures with light cream color.
3.3 Molecular identification of fungal cultures
The best matching sequences obtained in the GenBank database for the 84 representative isolates from ericaceous plants was summarized in Table 2. Phylogenetic analyses were also conducted on these sequences (Fig. 3).
Closest matches from FASTA searches between ITS sequences from endophytes isolated from root systems of different species of Ericaceae and known taxa from the Genbank and EMBL nucleotide databases.
| No. | Seq Num | Best blast match | Coverage | Similarity | Accession | Ericaceous host | Lineage |
| 1 | ER1M | Penicillium spinulosum | 100% | 100% | GU566191.1 | E. australis L. | Eurotiales |
| 2 | ER2M | Ericoid mycorrhizal sp. | 98% | 93% | AF072296.1 | C. vulgaris L. | Helotiales |
| 3 | ER3M | Mortierella sp | 100% | 99% | HQ608143.1 | E. umbellata L. | Mortierellales |
| 4 | ER4M | Umbelopsis sp | 97% | 100% | JQ912671.1 | E. umbellata L. | Mucorales |
| 5 | ER5M | Umbelopsis sp. | 97% | 100% | JQ912671.1 | E. umbellata L. | Mucorales |
| 6 | ER6M | Penicillium sp. | 100% | 99% | KF367517.1 | E. scoparia L. | Eurotiales |
| 7 | ER7M | Fusarium oxysporum | 99% | 100% | KU984712.1 | E. arborea L. | Hypocreales |
| 8 | ER8M | Fusarium oxysporum | 99% | 100% | KU984712.1 | E. arborea L. | Hypocreales |
| 9 | ER9M | Penicillium spinulosum | 100% | 100% | GU566191.1 | E. multiflora L. | Eurotiales |
| 10 | ER10M | Mortierella sp | 100% | 99% | LC127286.1 | E. multiflora L. | Mortierellales |
| 11 | ER11M | Penicillium sp. | 100% | 99% | JN798529.1 | V. corymbosum | Eurotiales |
| 12 | ER12M | Penicillium sp. | 100% | 99% | JN798529.1 | V. corymbosum | Eurotiales |
| 13 | ER13M | Eupenicillium sp | 100% | 97% | GU166451.1 | E. australis L. | Eurotiales |
| 14 | ER14M | Penicillium nodositatum | 100% | 99% | NR_103703.1 | E. umbellata L. | Eurotiales |
| 15 | ER15M | Fusarium oxysporum | 99% | 100% | KU984712.1 | A. unedo | Hypocreales |
| 16 | ER16M | Fusarium oxysporum | 99% | 100% | KU984712.1 | V. corymbosum | Hypocreales |
| 17 | ER17M | Pleosporales sp. | 89% | 91% | JX535184.1 | C. vulgaris L. | Pleosporales |
| 18 | ER18M | Alternaria sp | 100% | 100% | KX179491.1 | C. vulgaris L. | Pleosporales |
| 19 | ER19M | Fusarium oxysporum | 99% | 100% | KU984712.1 | E. arborea L. | Hypocreales |
| 20 | ER20M | Fusarium oxysporum | 99% | 100% | KU984712.1 | E. arborea L. | Hypocreales |
| 21 | ER21M | Penicillium nodositatum | 80% | 99% | NR_103703.1 | E.multiflora L. | Eurotiales |
| 22 | ER22M | Penicillium sp. | 100% | 99% | KF367517.1 | C. vulgaris L. | Eurotiales |
| 23 | ER23M | Penicillium nodositatum | 100% | 99% | NR_103703.1 | E. multiflora L. | Eurotiales |
| 24 | ER24M | Penicillium sp | 100% | 99% | KF367517.1 | E. multiflora L. | Eurotiales |
| 25 | ER25M | Mortierella sp | 100% | 99% | LC127286.1 | E. australis L. | Mortierellales |
| 26 | ER26M | Helotiales sp. | 93% | 96% | KR909148.1 | C. vulgaris L. | Helotiales |
| 27 | ER27M | Penicillium alexiae | 100% | 99% | NR_111869.1 | E. australis L. | Eurotiales |
| 28 | ER28M | Ericoid mycorrhizal sp. | 98% | 93% | AF072296.1 | E. umbellata L. | Helotiales |
| 29 | ER29M | Clad.sphaerospermum | 100% | 99% | KC311475.1 | C. vulgaris L. | Capnodiales |
| 30 | ER30M | Cladosporium sp. | 100% | 99% | LC133872.1 | A. unedo | Capnodiales |
| 31 | ER31M | Cystodendron sp. | 90% | 96% | EU434835.1 | C. vulgaris L. | Helotiales |
| 32 | ER32M | Coccomyces dentatus | 92% | 88% | GU138740.1 | C. vulgaris L. | Rhytismatales |
| 33 | ER33M | Penicillium sp. | 100% | 99% | KR812241.1 | A. unedo | Eurotiales |
| 34 | ER34M | Penicillium nodositatum | 80% | 99% | NR_103703.1 | E. multiflora L. | Eurotiales |
| 35 | ER35M | Pleosporales sp. | 89% | 91% | JX535184.1 | C. vulgaris L. | Pleosporales |
| 36 | ER36M | Helotiales sp. | 93% | 96% | KR909148.1 | C. vulgaris L. | Helotiales |
| 37 | ER37M | Helotiales sp. | 93% | 96% | KR909148.1 | C. vulgaris L. | Helotiales |
| 38 | ER38M | Penicillium nodositatum | 100% | 99% | NR_103703.1 | E. umbellata L. | Eurotiales |
| 39 | ER39M | Penicillium sp. | 100% | 99% | KC181935.1 | E. australis L. | Eurotiales |
| 40 | ER40M | Penicillium nodositatum | 100% | 99% | NR_103703.1 | E. umbellata L. | Eurotiales |
| 41 | ER41M | Ascomycota sp. | 100% | 86% | KU535786.1 | A. unedo | Unclassified ascomycota |
| 42 | ER42M | Penicillium nodositatum | 100% | 99% | NR_103703.1 | E.umbellata L. | Eurotiales |
| 43 | ER43M | Cystodendron sp. | 92% | 96% | EU434835.1 | C. vulgaris L. | Helotiales |
| 44 | ER44M | Fusarium oxysporum | 98% | 99% | KJ127284.1 | E.scoparia L. | Hypocreales |
| 45 | ER45M | Fusarium oxysporum | 98% | 99% | KJ127284.1 | E.arborea L. | Hypocreales |
| 46 | ER46M | Fusarium oxysporum | 98% | 100% | KJ127284.1 | A. unedo | Hypocreales |
| 47 | ER47M | Helotiales sp. | 93% | 96% | KR909148.1 | E.umbellata L. | Helotiales |
| 48 | ER48M | Fusarium oxysporum | 100% | 100% | KJ127284.1 | V. corymbosum | Hypocreales |
| 49 | ER49M | Fusarium oxysporum | 100% | 100% | KJ127284.1 | A. unedo | Hypocreales |
| 50 | ER50M | Ericoid endophyte sp. | 100% | 98% | AF252845.1 | C. vulgaris L. | Helotiales |
| 51 | ER51M | Ampelomyces sp. | 100% | 100% | AY148443.1 | A. unedo | Pleosporales |
| 52 | ER52M | Phialocephala fortinii | 100% | 99% | EU888625.1 | C. vulgaris L. | Helotiales |
| 53 | ER53M | Phialocephala fortinii | 100% | 99% | EU888625.1 | C. vulgaris L. | Helotiales |
| 54 | ER54M | Helotiales sp. | 93% | 96% | KR909148.1 | C. vulgaris L. | Helotiales |
| 55 | ER55M | Cryptosporiopsis brunnea | 100% | 99% | AF149074.2 | C. vulgaris L. | Helotiales |
| 56 | ER1F | Ericoid endophyte sp. | 97% | 98% | AF252848.1 | C. vulgaris L. | Helotiales |
| 57 | ER2F | Cystodendron sp. | 94% | 96% | EU434834.1 | C. vulgaris L. | Helotiales |
| 58 | ER3F | Helotiales sp. | 93% | 96% | KR909148.1 | C. vulgaris L. | Helotiales |
| 59 | ER4F | Helotiales sp. | 93% | 96% | KR909148.1 | E. umbellata L. | Helotiales |
| 60 | ER5F | Phialocephala sp. | 100% | 99% | AB847049.1 | V. myrtillus L. | Helotiales |
| 61 | ER6F | Phialocephala sp. | 100% | 99% | AB847049.1 | C. vulgaris L. | Helotiales |
| 62 | ER7F | Helotiales sp. | 93% | 96% | KR909148.1 | C. vulgaris L. | Helotiales |
| 63 | ER8F | Phialocephala sp. | 100% | 99% | AB847049.1 | C. vulgaris L. | Helotiales |
| 64 | ER9F | Phialocephala fortinii | 100% | 100% | FN678829.1 | C. vulgaris L. | Helotiales |
| 65 | ER10F | Phialocephala sp. | 100% | 99% | AB847049.1 | V. myrtillus L. | Helotiales |
| 66 | ER11F | Helotiales sp. | 93% | 96% | KR909148.1 | V. myrtillus L. | Helotiales |
| 67 | ER12F | Phialocephala cf. fortinii | 100% | 100% | FN678829.1 | V. myrtillus L. | Helotiales |
| 68 | ER13F | Phialocephala cf. fortinii | 100% | 100% | FN678829.1 | V. myrtillus L. | Helotiales |
| 69 | ER14F | Phialocephala subalpina | 100% | 99% | EF446148.1 | V. myrtillus L. | Helotiales |
| 70 | ER15F | Phialocephala fortinii | 100% | 100% | EU888625.1 | V. myrtillus L. | Helotiales |
| 71 | ER16F | Rhizodermea veluwensis. | 100% | 100% | KR859283.1 | C. vulgaris L. | Helotiales |
| 72 | ER17F | Ascomycota sp. | 93% | 96% | KC180744.1 | C. vulgaris L. | Unclassified ascomycota |
| 73 | ER18F | Helotiales sp. | 93% | 96% | KR909148.1 | V. myrtillus L. | Helotiales |
| 74 | ER20F | Helotiales sp. | 93% | 96% | KR909148.1 | V. myrtillus L. | Helotiales |
| 75 | ER21F | Phialocephala subalpina | 100% | 99% | EF446148.1 | V. myrtillus L. | Helotiales |
| 76 | ER22F | Cystodendron sp. | 92% | 96% | EU434835.1 | V. myrtillus L. | Helotiales |
| 77 | ER23F | Helotiales sp. | 93% | 96% | KR909148.1 | C. vulgaris L. | Helotiales |
| 78 | ER24F | Rhizodermea veluwensis. | 97% | 99% | KR859283.1 | V. myrtillus L. | Helotiales |
| 79 | ER25F | Phialocephala fortinii | 100% | 99% | EU888625.1 | V. myrtillus L. | Helotiales |
| 80 | ER26F | Phialocephala fortinii | 100% | 99% | AB671499.2 | C. vulgaris L. | Helotiales |
| 81 | ER27F | Phialocephala sp. | 100% | 99% | AB847049.1 | V. myrtillus L. | Helotiales |
| 82 | ER28F | Phialocephala subalpina | 100% | 99% | EF446148.1 | C. vulgaris L. | Helotiales |
| 83 | ER29F | Ericoid endophyte sp. | 96% | 98% | AF252845.1 | C. vulgaris L. | Helotiales |
| 84 | ER30F | Ericoid mycorrhizal sp. | 100% | 93% | AF072296.1 | V. myrtillus L. | Helotiales |
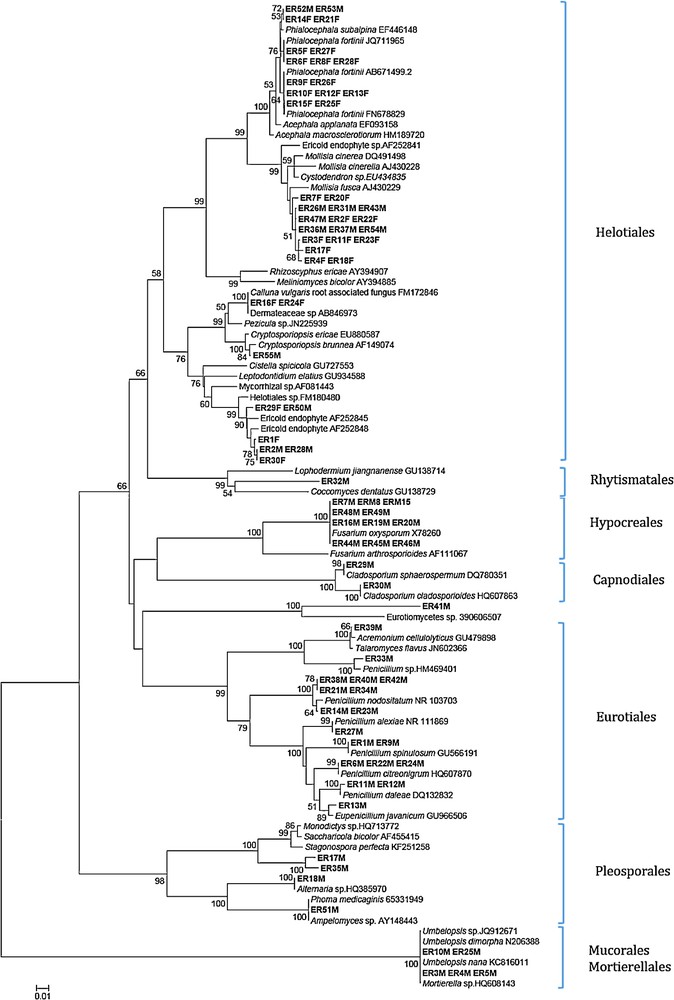
Neighbor-joining tree inferred from rDNA ITS sequences of Ericaceae root endophytic fungi and their closest GenBank matches (with accession numbers). Sequences from this study are in bold. Bootstrap support values (1000 replicates) are provided as percentage at the corresponding nodes when >50. Phylogenetic analysis was conducted in MEGA 4.0 [53] with the maximum composite likelihood method.
The most frequent fungal taxa isolated were ascomycetes (78/84 isolates) followed by zygomycetes (5/84 isolates). Ascomycetes were dominated by Helotiales (41/78 isolates) followed by Eurotiales (18/78 isolates), Hypocreales (11/78 isolates), Pleosporales (4/78 isolates), Capnodiales (2/78 isolates), Rhytismatales (1/78), whereas two ascomycetes isolates remained unidentified. Zygomycetes were represented by Mortierellales (3/5) and Mucorales (2/5).
Putative taxonomic affinities were assigned based on BLAST sequence similarity and the identities of the several most closely matched sequences obtained by BLAST (http://blast.ncbi.nim.nih.gov/Blast.cgi). Sequence analysis of cultured fungal ITS type has shown different types, the first group comprising significant portions of the isolated strains (ER52M, ER53M, ER14F, ER21F, ER5F, ER27F, ER6F, ER8F, ER28F, ER9F, ER26F, ER10F, ER12F, ER13F, ER15F, ER25F); these were most closely related to Phialocephala spp.
The second Helotiales group also comprised significant portions of the isolated stains (ER7F, ER20F, ER26M, ER47M, ER36M, ER37M, ER54M, ER3F, ER11F, ER23F, ER4F, ER18F), which were most closely related to various unidentified Helotiales species. Isolates ER2F, ER22F, ER31M and ER43M were designated as probable Cystodendron spp (96% similarity). Neighbor-joining analysis grouped isolates (ER29F, ER50M, ER1F, ER2M, ER28M, ER30F) (99% bootstrap) with different ericoid endophytes, Helotiales species (93% similarity) to ericoid mycorrhizal fungi.
A neighbor-joining analysis employing database sequences grouped these ER16F and ER24F (100% bootstrap) with C. vulgaris root associated fungus. The ER55M isolates matched (99% similarity) with Cryptosporiopsis brunnea and formed a strongly supported (100%) group with this species. The isolates belonging to Eurotiales, Hypocreales, Capnodiales, Pleosporales, Mucorales, and Mortierellales formed a strongly supported group.
3.4 Relationships between ericaceous fungal communities in the different sampling sites
3.4.1 Impact of ericaceous shrubs and isolation region on fungal distribution
Fungal isolates were grouped at the order level. Their presence in plants (frequency and percentage) according to the regions and the ericaceous species of isolation is displayed in Table 3. Statistical analysis confirmed the inequality of the proportions of the five regions (χ2 = 86 and P < 0.0001). Regarding the regions, most of the fungi were found in Melloussa (M) (42.31%) followed by Loge (L) (23.08%), whereas Sahel (S) (14.10%), Bab Berred (B) (11.54%), and Ourika (O)(8.97%) hosted less isolated fungi.
Frequency and percentage of the presence of fungal isolates in ericaceous plant as a function of regions and of ericaceous shrubs and fungi orders.
| Regions | Frequency | Percentage (%) |
| Bab Berred (B) | 27 | 11.54 |
| Ourika (O) | 21 | 8.97 |
| Melloussa (M) | 99 | 42.31 |
| Sahel (S) | 33 | 14.10 |
| Loge (L) | 54 | 23.08 |
| Ericaceous shrubs | ||
| Erica australis (4) | 15 | 6.41 |
| Erica arborea (3) | 24 | 10.26 |
| Erica multiflora (6) | 15 | 6.41 |
| Erica umbellata (7) | 18 | 7.69 |
| Vaccinium corymbosum (8) | 15 | 6.41 |
| Arbutus unedo (1) | 33 | 14.10 |
| Calluna vulgaris (2) | 75 | 32.05 |
| Erica scoparia (5) | 18 | 7.69 |
| Vaccinium myrtillus (9) | 21 | 8.97 |
| Fungi order | ||
| Eurotiales (eu) | 39 | 16.67 |
| Helotiales (he) | 108 | 46.15 |
| Mortierellales (mo) | 15 | 6.41 |
| Unidentified (un) | 15 | 6.41 |
| Rhytismatales (rh) | 3 | 1.28 |
| Mucorales (mu) | 12 | 5.13 |
| Hypocreales (hy) | 18 | 7.69 |
| Capnodiales (ca) | 15 | 6.41 |
| Pleosporales (pl) | 9 | 3.85 |
Besides, the fungi species were significantly found associated with C. vulgaris (32.05%), while they were least frequent on Vaccinium corymbosum (6.41%), E. australis (6.41%), and E. mutiflora (6.41%) (χ2 = 114.23 and P < 0.0001).
The most frequent fungal order was identified to be Helotiales (he) (46.15%) significantly associated with C. vulgaris (19.23%) and V. myrtillus (6.41%); it represented 16.67% in the Loge (L) and 15.38% in the Mellousa (M) regions. The least frequent order is Rhytismatales (rh) (1.28%). Again, the proportions were unequal, as evidenced by the Pearson chi-square test (χ2 = 320.53 and P < 0.0001).
Moreover, statistical analysis revealed an association between regions and ericaceous species, as shown by the Pearson Chi-square test of association (χ2 = 598.92; P < 0.0001).
To illustrate this, Vaccinium mytillus is fully specific to the Loge region (L; 100%) and E. scoparia is found only in the Sahel region (S; 100%). The association was observed to be strong, as evidenced by the Cramer coefficient of 0.8.
The same test showed association between regions and fungal orders; this was confirmed by the Pearson chi-square test of association (χ2 = 83.65 and P < 0.0001); this association was not significant (Cramer coefficient: 0.3). It was observed that only Rhytismatales (100%) are found in the Melloussa (M) region. Finally, the test showed an association between fungal orders and ericaceous species (χ2 = 181.53 and P < 0.0001); again this association was not strong enough, it gave a Cramer coefficient of 0.3. This is further supported by the 100% presence of the Rhytismatales order only in the root of C. vulgaris.
In conclusion, the Pearson chi-square test explained the presence and the degree of the association between regions, ericaceous host species, and fungal orders. However, the association was not enough strong, especially between regions and fungal orders, and between fungal orders and ericaceous species.
The frequency distribution of the fungi in the different host species or regions is shown in Tables A.2–A.5 (supplementary material).
3.4.2 Multiple correspondence analysis
As seen above, several associations were found between regions, ericaceous shrubs, and isolated fungi indicating that multiple correspondence analysis (MCA) can be performed to cluster all variables in distinct groups.
Main numerical characteristics of MCA are given in Table A.6 (supplementary material). The three first axes explained 84.7% of the variance in the dataset with the first axis explaining 33.8%, the second axis explaining 27.6% and the third axis explaining 23.3%.
The loadings of the 23 categories on the first factorial plan (axis 1–axis 2) are displayed in Fig. 4, and the 23 categories were clustered in three distinct groups.
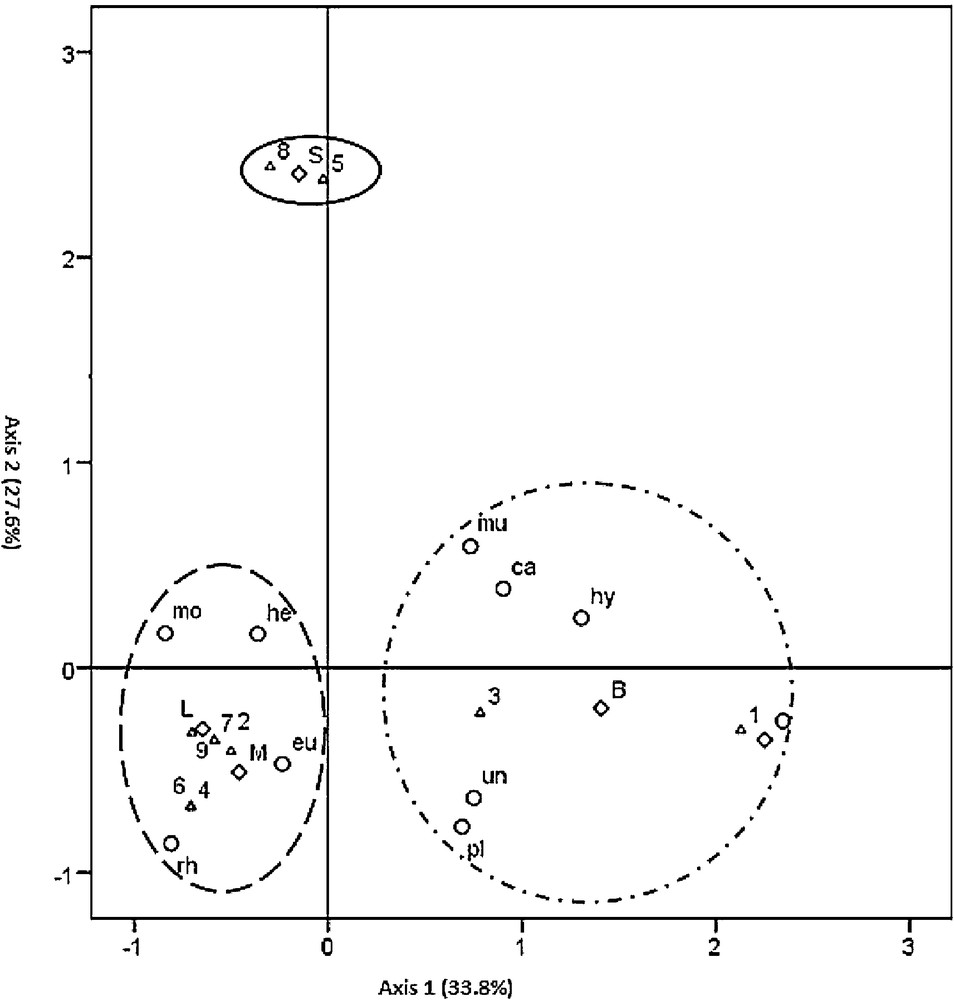
Multiple correspondence analysis (MCA) of the 23 categories of the three variables on the two first axes. ▵, Species; ○ Fungi-order; ♢ Region. B: Bab berred, O: Ourika, M: Melloussa, S: Sahel, L: Loge. 1: Arbutus unedo; 2: Calluna vulgaris; 3: Erica arborea; 4: Erica australis; 5: Erica scoparia; 6: Erica multiflora; 7: Erica embellata; 8: Vacinium corymbosum; 9: Vaccinium mytillus. eu: Eurotiales; he: Helotiales; mo: Mortierellales; mu: Mucorales; hy: Hypocreales; pl: Pleosporales; ca: Capnodiales; rh: Rhytismatales; un: unidentified.
Interestingly, axis 1 and axis 2 showed correlations with fungal assemblages, suggesting that host species and regions are involved in structuring fungal assemblages:
- • the first group (G1) corresponds to the association of the Bab Berred region (B) and Ourika (O) with the group of Capnodiales (ca), Hypocreales (hy), and Mucorales (mu) fungal orders, pleosporales (pl) and unidentified fungi (un) with E. arborea (3) and A. unedo (1);
- • the second group (G2) associated the Sahel region (S) with E. scoparia (5) and V. corymbosum (8) ericaceous shrubs; this group seemed not to be associated with any specific fungal order;
- • the third group (G3) was clearly distinct from the two other groups. It associated the Loge (L) and Melloussa (M) regions with the Helotiales (he); and Rhytismatales (rh) fungal families with C. vulgaris (2), V. myrtillus (9), E. australis (4), E. multiflora (6), and E. umbellata (7).
4 Discussion
4.1 Diversity of fungal species associated with Ericaceae in Mediterranean contrasting ecosystems
The diversity of fungal endophytes in the roots of Ericacea taxa has been reported previously [3,9–12,17,56–58]. However, the diversity of Ericaceae root endophyte fungi is relatively low compared to arbuscular mycorrhizal and ectomycorrhizal plants [33]. Besides, studies concerning this topic are missing in some regions, especially in the Mediterranean ones. These environments can offer an interesting opportunity for the study of fungal diversity in ericaceous plants [59] as they are largely unexplored. This research represents the first attempt to isolate and study the diversity of endophytes present in the root system of nine ericaceous species grown in specific areas in Morocco and France.
In this study, we used both cultural methods and DNA analysis of isolated fungi to identify the endophytes obtained. This approach has been adopted over the last years to identify sterile endophytic mycelia by many authors [60–63]. The result indeed has shown a large diversity of fungi isolates belonging to Ascomycetes. Helotiales isolates were the most dominant; this emphasizes their importance inside the fungal communities associated with the Mediterranean Ericaceae. Interestingly, our findings were similar to those reported by Tedersoo et al. [64], who targeted ascomycetous communities associated with the ectomycorrhizal roots of various hosts in Tasmani and those of Walker et al. [11] in the Arctic tundra.
The sequencing of the studied isolates revealed that the most common isolates belong to the Phialocephala–Acephala complex (PAC), representing 50% in total of the Helotiales. The sequence analysis confirmed some isolates as P. fortinii (99–100% similarity), suggesting that this taxon or its sibling species might be the dominant root entophytes of ericaceous species in the sites (M), (S), and (L). However, no Phialocephala spp strains were isolated from site (O) and site (B). The plant communities of the sites (S) and (L) were some pine and mixed forest; ericaceous shrubs were in site (M), while A. unedo was found in sites (O) and (B). Moreover, the sites (M), (S) and (L) are located in the North, with mostly sub-humid climate, relatively high precipitation and lower pH, than that at site (O) situated in the south of Morocco, with less precipitation in the semi-arid climate. It seems that the abundance of the Phialocephala spp. may be related to prevailing plant communities and edaphic factors [57,65,66].
DSE colonization is characterized by the formation of microsleclerotia in the host root. Nonetheless, a few DSE species were reported to form intra-radical structures resembling those formed in mycorrhizal symbioses [67]. The studies on the functional aspects of these intra-radical hyphal structures, i.e. nutrient transfer and/or plant growth response to colonization, are few [18,36] in our context; further investigations into these groups of fungi are needed due to their dominance and possible functional importance to ericaceous plants.
Besides PAC, the screened ericaceous roots hairs hosted relatively diverse spectrums of mycobionts, for example, the ITS sequence analyses have shown the presence of Cryptosporiopsis species at a low frequency. Cryptosporiopsis spp are known root-inhabiting fungi, and they colonize Ericaceae roots as an endophyte [68]. Related taxa of Cryptosporiopsis (C. ericae and C. brunnea) were isolated from some Ericaceae plants, such as Vaccinium ovalifolium, Vaccinium membranaceum, and Gaultheria shallon [69]. Moreover, Chambers et al. [70] have shown that an isolate of Cryptosporiopsis species formed dense ERM-like coils in occasional cells in Woollsia pungens root hairs. However, Zhang et al. [57] isolated C. ericae assemblages from Rhododendrons and have confirmed their ericoid mycorrhizal status; in this study, additional research is needed to elucidate the functional status of this species.
A neighbor-joining analysis employing database sequences grouped ITS types with 12 isolates; these were designated as the Helotiales species. Two isolates were grouped with different unidentified C. vulgaris root associated fungus. Surprisingly, six isolates were grouped together with different unidentified ericoid endophyte fungi from C. vulgaris at contrasting field sites [57]. ITS sequence analysis showed that the isolates have a high affinity for root endophytes from C. vulgaris and are probably homologous fungi.
In contrast, the study was not able to obtain any isolate belonging to the ErM fungus Rhizoscyphus ericae, which is prominent in most studies on Ericaceae. This result was not significant because most of the screened root hairs contained ericoid mycorrhizae [71]. This might be explained by their relatively slower growth on artificial isolation media, especially when the fast-growing DSE dominate the root-associated fungal communities [11,19]. To prove the presence of intracellular hyphal structures to confirm the putative ericoid mycorrhizal status, especially from areas, which have not yet been investigated, the investigation had to be performed on cultivation-based methods, followed by re-synthesis and nutrient transfer experiments [33].
The study finally revealed the presence of common soil saprobic/parasitic fungi known to associate with ericaceous roots, especially in plants from Morocco. This could be expected as the same saprobic/parasitic community had been reported by Bruzone et al. [19] in association with ericaceous shrubs.
4.2 Impact of region and plant hosts on ericaceous fungal communities’ structures
The total diversity of Ericaceae mycobionts was relatively high, but the most abundant one was the Helotiales order, dominated by Phialocephala spp., Helotiales spp and unidentified ericoid fungi, which accounted for 46.15% of the total mycobionts selected. They showed a strong preference for certain Mediterranean sites, characterized by hot, dry summers, and cool, wet winters, under humid bioclimates, such as Mellousa (15.38%), La Loge (16.67%), whereas they were less abundant in other sites such as Ourika (1.28%). The Helotiales showed as well a strong preference for C. vulgaris (19.23%) and V. myrtillus (6.46%), our finding is in agreement with previous studies [20,35,72], where P. fortinii have been detected as an associate of C. vulgaris roots.
Statistical analysis has shown an association between regions, fungal orders, and ericaceous species. Surprisingly, this association was not strong enough to conclude that there is significant influence of both plant hosts and regional factors on associated fungal communities.
Previous studies carried out by Kjoller et al. [43] and Walker et al. [11] targeted common co-occurring Ericaceae in sub-Arctic mire and Arctic tundra habitat (respectively). They provided no support for host preference and showed that the host may not be an important driver for the composition of root fungal communities in the Arctic Ericaceae. On the contrary, Kernaghan [73] suggested that mycorrhizal diversity is controlled by many factors, among them the host plant. Ishida and Nordin [47] observed distinct communities in V. myrtillus and V. vitis-idaea in boreal forest stands dominated by Norway spruce; Bougoure et al. [20] also reported distinct fungal communities in V. myrtillus and C. vulgaris in pine forest sites in Scotland. Both views suggest the influence of plant hosts, as a driver of fungal communities structures might thus be dependent on the region studied.
The success of ericaceous plants in ecosystems is the result of the ability of the plant/fungal symbiosis to succeed in conditions with extreme low levels of mineral N and P and high levels of recalcitrant organic matter. In this context, other studies have proved that plant diversity is maintained by their capability to acquire N from different organic forms [74]; the same results were reported by Kjoller et al. [43] and Walker et al. [11]. Subsequently, the differences in N use varied with species [18] rather than between species [14].
Besides, Sun et al. [40] targeted ericoid mycorrhizal fungi and other fungal assemblages in the roots of Rhododendron decorum in the Southwest of China; they concluded that the ericoid mycorrhizal (ErM) and non-mycorrhizal (NEM) fungi are affected by different factors; the host's genetic composition is more important for ErM while geographic factors are more important for NEM assemblages.
Through these studies, the influence of hosts in controlling the community assembly of root-associated fungi is still under debate and need more research to determine the different mechanisms responsible for the maintenance of this diversity; this emphasizes the need to study the different factors that could affect fungal communities in a Mediterranean context.
5 Conclusion
The investigation of ericaceous endophytes colonizers of a variety of healthy Ericaceae in Mediterranean ecosystems has revealed a large diversity of fungi. These were dominated by ascomycetes, with taxa closely related to Dark Septate Endophytes (DSE), unidentified ericoid endophyte fungi. The analyses suggest that a number of associations exist between the Helotiales and Ericaceae; however, these associations are not strong enough, suggesting that other factors may be affecting the diversity of fungal communities of ericaceous shrubs and should be explored.
The isolation of beneficial Helotialean endophytes from ericaceous roots encourages and permits to carry out re-synthesis experiments and to evaluate nutrient transfer systems to resolve the ability of some putative ericoid mycorrhizal strains obtained to form mycorrhizae symbiosis and improve the growth of other domesticated ericaceous species such as Vaccinium spp.
Acknowledgements
This study was financially supported by the “Programme de recherche agronomique pour le développement” (PHC PRAD No. 28044TM). The authors thank Dr. Ibn Tattou and Hamid El Khamer from the Scientific Institute in Rabat for their help in the identification of ericaceous species in different locations in Morocco.


