1 Introduction
The Indian aromatic rice, often called “Basmati” is nature's gift to the Indian sub-continent and human kind at large. Basmati rice is highly priced in the domestic as well as international markets. Traditional varieties of Basmati rice are durable, photosensitive and susceptible to all rice diseases prevalent in the area. These varieties are also tall with weak stem and therefore they lodge under high-input agriculture, resulting in yield loss and inferior quality. Therefore, the development of short-duration, dwarf, photo-insensitive, and disease-resistant varieties of Basmati rice requires special considerations. Basmati rice cultivated in the northwestern part of the Indo-Gangetic plains of the Indian subcontinent is highly valued on the international market due to its unique combination of aroma, grain, cooking quality attributes and eating qualities such as fluffy texture of cooked rice and high volume expansion during cooking, which is characterized by linear kernel elongation with minimum breadth wise swelling, palatability, easy digestibility, and long shelf life [1].
Ranbir Basmati grown in Ranbir Singh Pura (R. S. Pura) areas of Jammu region in Jammu and Kashmir, India is one of the best traditional Basmati varieties of the country. It possesses long, slender, translucent grains with strong and pleasant aroma. It is a short-duration variety that matures 20–30 days early to Basmati 370 while maintaining strong aroma and excellent cooking quality. However, the tall stature and thin stems of Ranbir Basmati render the variety prone to lodging and is also susceptible to BB disease, because of which it is losing its popularity among farmers in the state.
Indian traditional Basmati rice suffers from a severe problem of lodging that arises mainly due to its tall height, weak stem, and uncontrolled use of nitrogen fertilizers. Plants with tall height are prone to lodging as the stems of these plants are weak to support the heavy grain of the high-yielding varieties, which ultimately leads to low grain–straw ratio or low harvest index. The semi-dwarf gene (sd1) controls the semi-dwarf phenotype in rice and is introgressed in many modern rice cultivars [2]. It was introduced in most indica rice varieties in 1960s in IR8, the cornerstone of Green Revolution. The presence of sd1 can be only determined from the final height of the plant, which is scored late in development and is influenced by environmental factors. Furthermore, sd1 is a recessive gene and heterozygosity at the sd1 locus can be determined only by progeny testing. The development of a molecular marker for sd1 would therefore provide breeders with an additional option to test the presence of dwarf or tall alleles rapidly at any growth stage of the plant. A number of varieties has been developed by using the semi-dwarf (sd1) gene that is originated from the Chinese cultivar “Dee-geo-woo-gen” (DGWG) and is known to encode GA20 oxidase-2 (GA20ox-2) that controls gibberellins’ (GA) biosynthesis pathway [2–4]. However, the development of molecular marker for sd1 gene has now provided a way to determine the presence of short or tall plant at any growth stage.
Another important constraint that confines the rice production is the diseases caused by necrotrophic agents, and one of them is bacterial blight disease caused by gram negative proteobacterium Xanthomonas oryzae pv. oryzae, which is the major threat to rice production and makes grain quality declines by up to 80% in India [5]. The most economic and environmentally sustainable way to overcome the disease is to develop resistant varieties and, so far, more than 41 BB resistant genes have been identified in rice, of which 8 have been cloned and characterized [6]. Among the BB resistance genes, Xa21 gene is the dominant gene conferring broad spectrum resistance against various BB virulent pathogens [7]. It is a member of a small multigene family with seven members and they are linked together, suggesting that Xa21 gene is part of a complex locus [8]. Xa21 gene is mapped on chromosome 11 and encodes receptor-like kinases. Specific markers are available for Xa21 gene, which has been introgressed into several elite rice varieties and hybrid rice parental lines in combination with other dominant or recessive genes for providing durable resistance against BB [9,10]. Another major resistance gene, xa13, identified from rice variety BJ1 and mapped on the long arm of rice chromosome 8, has been introgressed into various elite varieties of rice along with the Xa21 gene [7,11,12].
Both Xa21 and xa13 are broad-spectrum R genes for bacterial blight resistance. The two R genes have different resistance spectrum, but provide durable resistance against the disease. NILs carrying both genes (Xa21 and xa13) were more resistant to BB disease compared to NIL's carrying single genes (either Xa21 or xa13). This may be due to lines carrying single resistance genes resulting in the breakdown of resistance [13]. Hence, pyramiding of multiple resistance genes is widely used for introgression of desirable traits to achieve the enhanced resistance to specific disease. The enhanced level of resistance could be the result of synergistic action or quantitative complementation between these resistant genes [14].
Pyramiding of resistance genes using conventional breeding is difficult to achieve due to several reasons, including epistatic effects of genes controlling resistance and the non-availability of screening facilities for multiple biotic stresses; in addition, screening is restricted only to specific seasons. Molecular markers can hasten the resistance-breeding efforts [7]. The segregating plants can be selected by target molecular markers instead of its phenotype, and introgression of multiple resistance genes can be easily monitored in the population. The marker-assisted selection (MAS) scheme for pyramiding important genes encompasses a rapid background recovery of the recurrent parents and maintains the exquisite quality characteristics of rice, which could be an effective approach for rice improvement programs [7,15].
Keeping in view the importance Ranbir Basmati among the traditional Basmati varieties of the Jammu region in the State of Jammu & Kashmir and the severity and significance of damage caused by the disease and lodging have necessitated the development of effective strategies for their management. The present work was undertaken with the aim to introgress two BB resistance genes (xa13 and Xa21) along with semi-dwarf gene (sd1) from PAU148 (Basmati introgressed line xa13, Xa21 and sd1 genes) into Ranbir Basmati. The identified introgressed line having all the target genes combination will provide broad resistance to Xoo strains and significant agro-morphologically superiority in comparison to Ranbir Basmati, which can be further evaluated for release of a variety and or used in breeding programmes.
2 Materials and methods
2.1 Plant material and crossing plan
Ranbir Basmati possessing strong aroma and excellent cooking quality was used as a recurrent parent while a PAU148 possessing BB (xa13 and Xa21) resistant genes along with semi-dwarf (sd1) gene was used as a donor parent. The marker-assisted backcross breeding (MABB) scheme was used to pyramid semi-dwarf and BB resistance genes in Ranbir Basmati. The individual F1 plants produced by crossing Ranbir Basmati/PAU148 and confirmed by gene-specific markers were backcrossed with recurrent parent to generate the backcross population. The backcross method was followed up to BC2F2 generation and at each generation, foreground selection as well as background selection was performed to select positive plants carrying desirable genes. The schematic diagram for pyramiding of semi-dwarf and BB resistance genes in the recurrent parent is depicted in Fig. 1. Further, before using the donor parent PAU148 for introgression of BB resistance and semi-dwarf genes in Ranbir Basmati, validation of the molecular markers linked to these genes was carried out in the selected donor parent. The details of the molecular markers (xa13, Xa21 and sd1) are presented in Table 1.
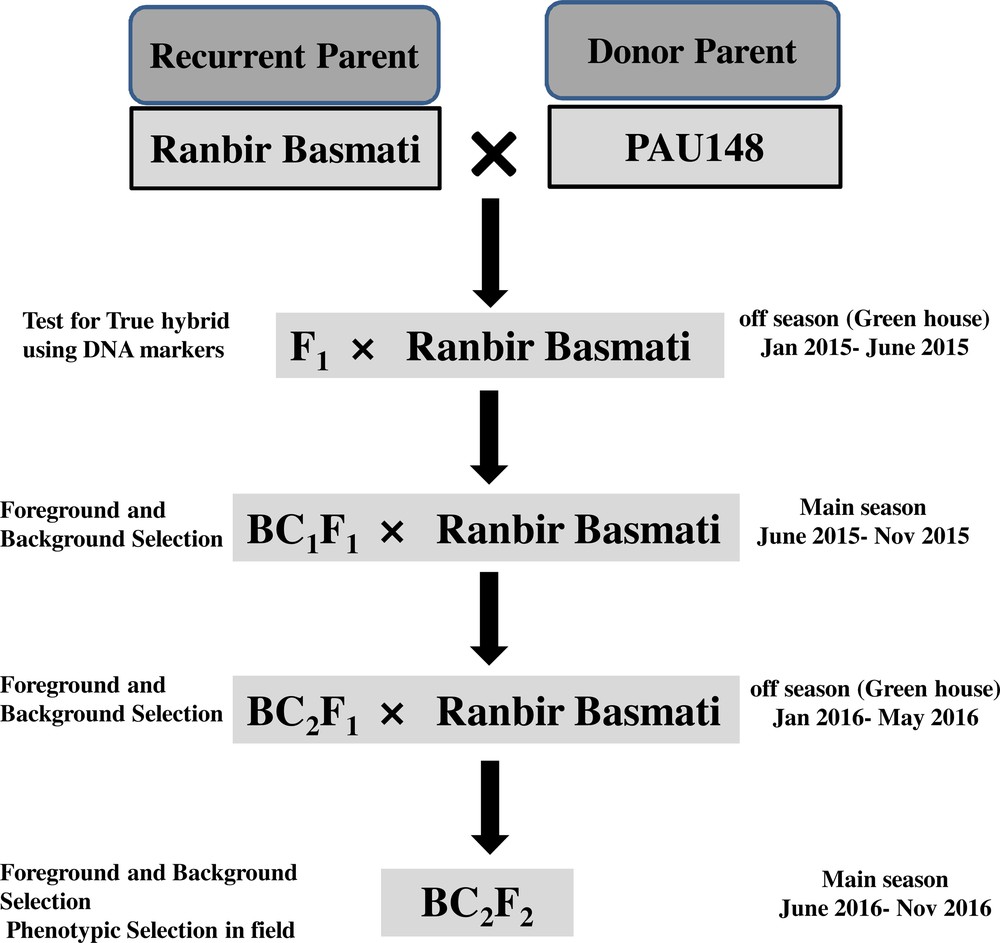
The schematic diagram of the crossing program for the introgression of semi-dwarf and bacterial blight (BB) resistance genes in the recurrent parent. BCF represents the backcross filial population.
Gene specific markers used for foreground selection of BB resistance and semi-dwarf genes in marker-assisted backcross breeding.
| Diseases/stresses | Linked genes | Markers | Chromosome number | Primer sequence | Band type | References |
| Bacterial Blight |
xa13 (3.8 cM) |
xa13prom | 8 | F:GGCCATGGCTCAGTGTTTAT R:GAGCTCCAGATCTCCAAATG |
STS | (Sundaram et al., 2008) |
| Xa21 (0.2 cM) |
pTA248 | 11 | F:AGACGCGGAAGGGTGGTTCCCGG R:AGACCGGTAATCGAAAGATGAAA |
STS | (Ronald et al. 1992) | |
| Semi-dwarf | sd1 | ‘h’ | 1 | F:CACGCACGGGTTCTTCCAGGTG R:AGGAGAATAGGAGATGGTTTACC |
STS | (Monna et al., 2002) |
2.2 DNA isolation and PCR amplification
Total genomic DNA was extracted from rice leaves following the modified protocol of Sahu et al. [16] and quantified spectrophotometrically. The PCR reaction mixture for foreground selection of xa13 and Xa21 contained 50 ng genomic DNA template, 10 pmoL of each of the primers, 5 mM dNTP's, 10X PCR buffer, 1 U taq polymerase in a volume of 10 μL. For sd1 gene, the PCR reaction mixture contained 5X Q-solution, 10 mM dNTP's, 2.5 U Hot star taq polymerse. The PCR profile for all the genes studied was different with respect of temperature and timing and is provided in Supplementary Table 1. The PCR amplified products were separated by electrophoresis on a 2% agarose gel and visualized on gel documentation system (Bio-Rad Laboratories Inc., USA).
2.3 Survey of parental polymorphism between parental lines using SSR markers
A parental polymorphism survey was conducted with the recipient parent (Ranbir Basmati) and donor parent (PAU148), and a total of 384 SSR markers selected from www.gramene.org were screened for their polymorphism. The SSR markers were selected from each short and long arm of rice chromosome 12. Out of 384 SSR markers, 51 polymorphic markers uniformly spanning across the genome were selected (Supplementary Table 2). These polymorphic SSR markers were used for background selection in order to select plants having maximum recovery of the recurrent parent genome. Graphical GenoTypes (GGT) version 2.0 [17] software was used for the assessment of the genomic contribution of the parent in the selected genotypes based on SSR marker data. Further, polymorphic SSR primers were resolved on 3.5% agarose gel and visualized.
2.4 Screening for resistance to bacterial blight (BB)
The BC2F2 introgressed lines carrying xa13 and Xa21 genes both either single or in combination were selected and evaluated for BB resistance. Leaf clip inoculation method was used for artificial inoculation of pyramided lines with BB disease [18]. The inoculum was prepared by suspending the bacterial mass in distilled water to a concentration of 106 cells/mL. The inoculation was carried out by clipping the tip of the leaf at the tillering stage with scissors that had been dipped into the inoculum. The symptoms become visible after five to six days after inoculation and scoring was done using IRRI-SES scale [19] after 15 days of inoculation (Supplementary Table 3).
2.5 Characterization for agro-morphological and grain quality traits
The pyramided lines along with parents (Ranbir Basmati and PAU148) were evaluated at Experimental Research Farm, Sher-e-Kashmir University of Agricultural Sciences and Technology of Jammu (SKUAST-J) during kharif season in 2016. Twenty-five day-old seedlings of selected pyramided lines were transplanted with spacing of 15 × 20 cm in a randomized complete block design (RCBD) with two replications. Data were collected from five randomly selected plants from each entry in each replication for agro-morphological and grain quality characters including plant height, days to 50% flowering, days to maturity, effective tillers per plant, panicle length, 1000 grain weight, yield per plant, grain length, grain breadth, L/B ratio, kernel length after cooking (KLAC), kernel breadth after cooking (KLBC) and aroma. The scale used for aroma test was 0, 1, 2, 3 for no, mild, strong, and very strong aroma, respectively [20].
2.6 Cluster analysis
The SSR–PCR bands were examined under ultra violet transilluminator and photographed under gel documentation unit (Bio-Rad Laboratories Inc., USA). The SSR bands were counted and scored manually as 1 for their presence and 0 for their absence to generate the binary data for diversity analysis. The molecular data was analysed using NTSYS-PC (Numerical Taxonomy and Multivariate Analysis System) computer package [21]. The genetic similarity between accessions was calculated by Dice (SSR) and Jaccard's similarity coefficient. The dendogram was constructed based on sequential UPGMA (unweighted pair group method with arithmetic mean) using software package NTSYS PC 2.11 to infer genetic relationships and phylogeny.
3 Results
3.1 Validation of markers linked to the resistance genes xa13, Xa21 and sd1 and identification of parental polymorphic markers
The donor parent (PAU148) was validated for the presence of xa13, Xa21 and sd1 genes using gene-specific markers xa13 promo, pTA248 and h, respectively. The amplified fragment with respect to xa13 promo in resistant and susceptible lines was 500 bp and 250 bp [7]. With primer pair pTA248, it was 1000 bp and 650 bp [8] and, with respect to primer pair ‘h’, it was 348 bp and 731 bp, respectively [22] (Fig. 2). The results revealed that all the markers used for xa13, Xa21 and sd1 genes were able to distinguish resistant lines from susceptible as well as from dwarf to tall ones.
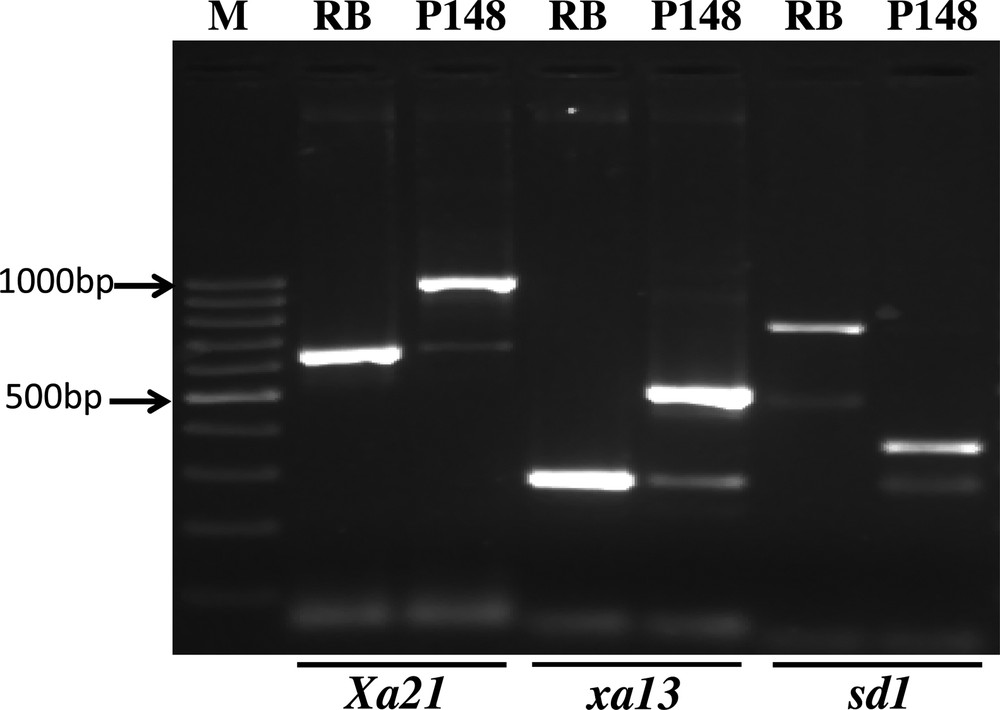
Validation of the markers linked to the resistance genes. The amplified fragments with respect to xa13 promo in resistant lines and susceptible lines were 500 bp and 250 bp, respectively. The primer pair pTA248 amplified fragments in the resistant line and the susceptible line were 1000 bp and 650 bp, respectively. With respect to primer pair ‘h’, the amplified fragment in the donor parent and the recurrent parent were 348 bp and 731 bp, respectively.
Parental polymorphism survey was carried out using 384 SSR markers distributed across the 12 chromosomes of rice. Out of 384 SSR markers, 51 (13.28%) markers were found to be polymorphic between Ranbir Basmati and PAU148, while 333 (86.71%) markers were monomorphic. The 51 polymorphic SSR markers were used for background selection at each generation. The number of alleles varied from 1 to 2. The Polymorphism Information Content (PIC) value was calculated by using the formulae given by Roldan-Ruiz et al. [23]. The highest PIC value was shown in case of primer RM18 and RM22 (0.48), while the lowest PIC value was observed in RM317, RM499, RM3625, and RM527 (0.07) (Supplementary Table 2).
3.2 Pyramiding of semi-dwarf (sd1) and bacterial blight (xa13 and Xa21) resistance genes
Marker-assisted backcross breeding (MABB) method was used to introgress semi-dwarf and BB resistance genes in Ranbir Basmati. During each generation from F1 to BC2F2, foreground selection was carried out and plants having resistance alleles of all the three genes (xa13, Xa21 and sd1) were selected and advanced to the next generation. F1 plants were also tested for the hybridity and true F1S were backcrossed with recipient parent to get BC1F1 generation seeds. In BC1F1, a total of 325 plants were grown and screened for the presence of BB resistance and semi-dwarf genes using molecular marker xa13-promo, pTA248, and ‘h’. Out of 325 BC1F1 plants, 36 plants were found to be positive for xa13 + sd1 genes, 13 for xa13 + Xa21 and 23 for Xa21 + sd1 genes. Totals of 26, 60 and 82 plants showed positive for xa13, Xa21 and sd1 genes, respectively. A total of 17 plants were found to be positive for all three genes, viz. xa13, Xa21 and sd1.
In subsequent BC2F1 generation out of 150 plants, 5 plants showed positive for all three genes (xa13, Xa21 and sd1). Thirty-five plants were positive for xa13 + sd1, 17 for Xa21 + sd1 genes and 16 for xa13 + Xa21 genes. Similarly, 29 plants were positive for xa13, 14 for Xa21 and 37 for sd1.
Furthermore, in BC2F2 generation, out of 121 plants, 19 plants were found to be positive for all three genes (xa13, Xa21 and sd1), while 34 plants were found to be positive for xa13 + sd1 genes, 25 for Xa21 + sd1 genes and 23 for Xa21 + xa13 genes. For Xa21, xa13 and sd1 genes 34, 44 and 93 plants, respectively, were found to be positive (Fig. 3). Background selection was started from BC1F1 and continued up to BC2F2 generation. In each generation, plants showing maximum recovery of recurrent parent were advanced to the next generation. Further, the background selection of 19 introgressed line (Fig. 4) resulted in the highest recurrent parent genome (RPG) recovery in introgressed line SBTIL121 (86.9%) (Fig. 5). The superior BC2F2 plant was further advanced to the next generation.
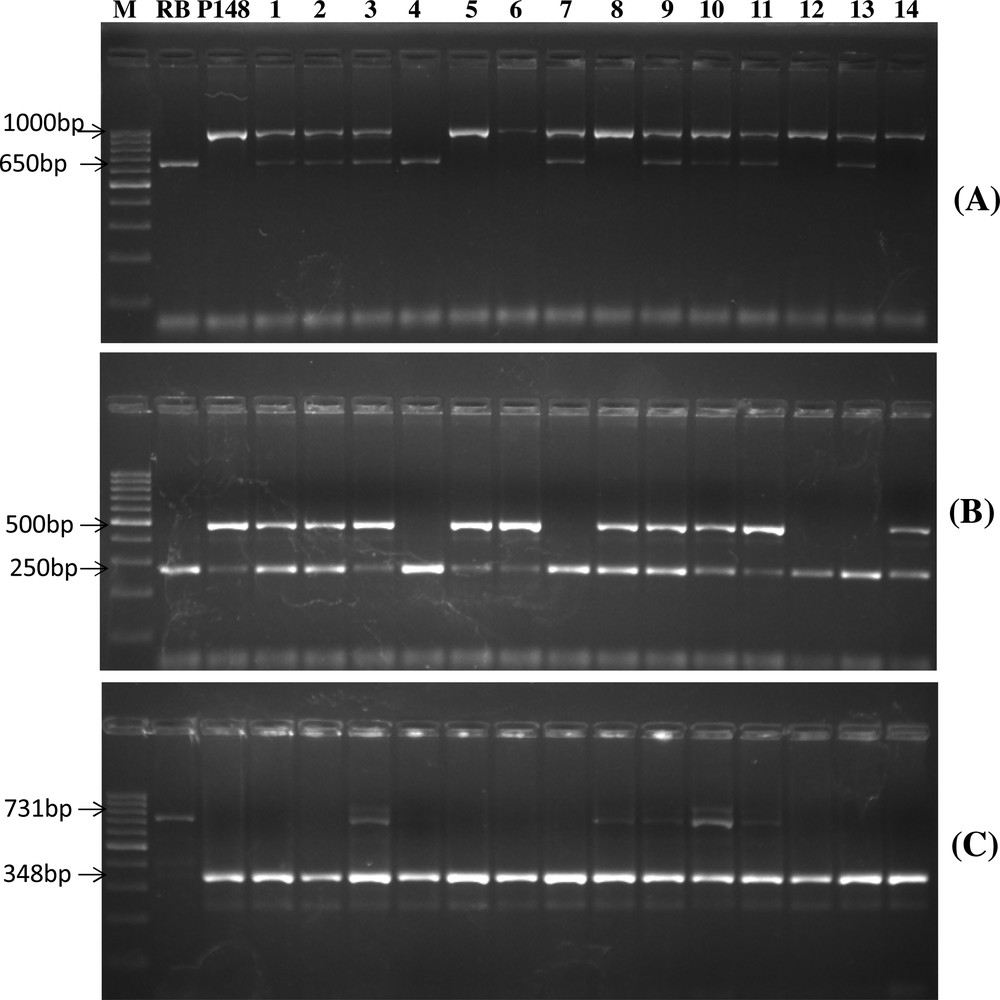
Foreground selection of BC2F2 population A) for Xa21gene B) for xa13 gene C) for sd1 gene. The Plant number 1, 2, 3, 5, 6, 8, 9, 10, 11 and 14 are positive for all three genes. M, 100 bp ladder, P148, PAU148, RB, Ranbir Basmati.
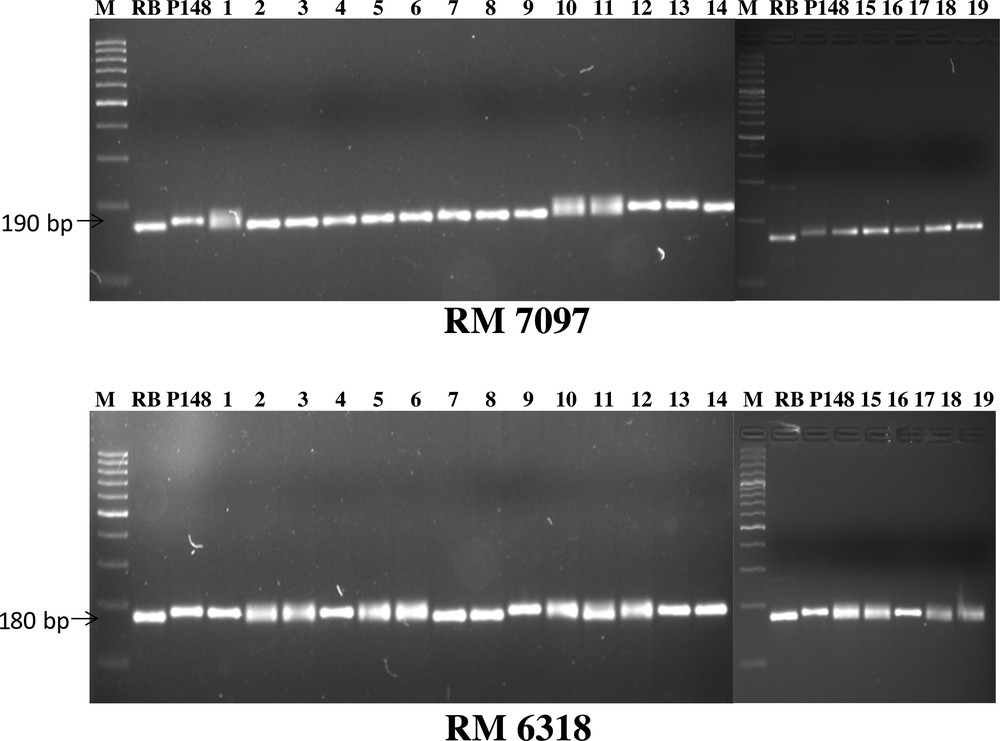
Background selection of BC2F2 positive population using SSR marker RM 7097 and RM 6318 (M, 100 bp ladder, P14, PAU148, RB, Ranbir Basmati).
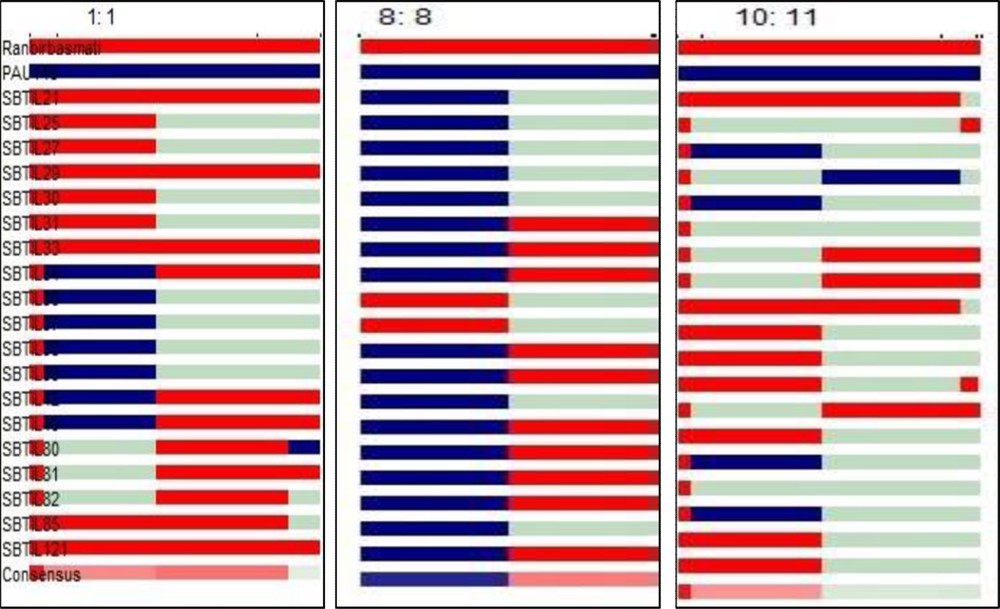
Analysis of the genome introgression of 19 introgressed lines associated with resistance genes using software Graphical Geno Types (GGT) (Van Berloo 1999). 1.1 indicates sd1 on chromosome 1; 8.8 indicates xa13 on chromosome 8; 10.11 indicates Xa21 on chromosome 11 of introgressed lines.
3.3 Screening of the BC2F2 population for bacterial blight resistance
The NILs along with parents were screened for BB resistance. The Xoo strain, which was most prevalent in the area, was used as an inoculum. The donor parent PAU148 showed lesion length between 1–5% with disease scoring scale value ‘1′, while the recurrent parent Ranbir Basmati showed average lesion length between 51–100% with disease score ‘7′. The pyramided lines showed very small lesion length (disease score 1) and were highly resistant to Xoo isolates. The introgressed line SBTIL121 having the maximum recovery of recurrent genome and possessing the superior grain quality displayed resistance to BB with less lesion length (Fig. 6). The plants with a score of 1 were considered as resistant, 3 as moderately resistant, while those with 7–9 were considered as susceptible. The IRRI standard evaluation system (IRRI–SES scale) was adopted for the screening of the introgressed lines [19]. Twenty-three introgressed lines carrying two genes together in homozygous condition (xa13xa13 Xa21Xa21) showed lesion lengths between 1 and 5% with disease score ‘1′, while it is significant to note that single BB-resistant genes in homozygous condition (i.e. either xa13xa13 or Xa21Xa21) showed lesion lengths between 6 and 12% with disease score ‘3′. The single BB-resistance gene showed partial level of BB resistance (Table 2). Thus, all the lines having xa13 + Xa21 gene combination showed a higher level of resistance, while lines showing either xa13 or Xa21 alone showed moderate levels of resistance against Xoo isolates.

Leaves of BC2F2 introgressed line SBIL121 showing their resistance to bacterial blight compared to those of Ranbir Basmati (recurrent parent) and PAU148 (donor parent).
Disease scoring of the introgressed lines along with parents (scale 0–9).
| Genotypes | Gene Combination | Disease scoring | Resistant/Susceptible | Genotypes | Gene Combination | Disease scoring | Resistant/Susceptible |
| Ranbir Basmati | – | 7 | Susceptible | SBTIL 38 | xa13 + Xa21 | 1 | Resistant |
| PAU148 | xa13 + Xa21 | 1 | Resistant | SBTIL 39 | xa13 + Xa21 | 1 | Resistant |
| SBTIL05 | xa13 | 3 | Moderately resistant | SBTIL 40 | Xa21 | 3 | Moderately Resistant |
| SBTIL06 | xa13 | 3 | Moderately resistant | SBTIL 41 | Xa21 | 3 | Moderately Resistant |
| SBTIL07 | xa13 | 3 | Moderately resistant | SBTIL 42 | xa13 + Xa21 | 1 | Resistant |
| SBTIL08 | xa13 | 3 | Moderately resistant | SBTIL 44 | xa13 | 3 | Moderately Resistant |
| SBTIL09 | xa13 | 3 | Moderately resistant | SBTIL 45 | xa13 | 3 | Moderately Resistant |
| SBTIL12 | xa13 | 3 | Moderately resistant | SBTIL 46 | xa13 | 3 | Moderately Resistant |
| SBTIL14 | xa13 | 3 | Moderately resistant | SBTIL 47 | Xa21 | 3 | Moderately Resistant |
| SBTIL15 | xa13 | 3 | Moderately resistant | SBTIL 48 | xa13 + Xa21 | 1 | Resistant |
| SBTIL 16 | xa13 | 3 | Moderately resistant | SBTIL 49 | xa13 + Xa21 | 1 | Resistant |
| SBTIL 17 | Xa21 | 3 | Moderately Resistant | SBTIL 51 | xa13 | 3 | Moderately Resistant |
| SBTIL 18 | Xa21 | 3 | Moderately resistant | SBTIL 52 | xa13 | 3 | Moderately Resistant |
| SBTIL 19 | Xa21 | 3 | Moderately resistant | SBTIL 53 | xa13 | 3 | Moderately Resistant |
| SBTIL 20 | xa13 + Xa21 | 1 | Resistant | SBTIL 57 | xa13 | 3 | Moderately Resistant |
| SBTIL 21 | xa13 + Xa21 | 1 | Resistant | SBTIL 58 | xa13 | 3 | Moderately Resistant |
| SBTIL 22 | xa13 | 3 | Moderately resistant | SBTIL 78 | Xa21 | 3 | Moderately Resistant |
| SBTIL 23 | xa13 | 3 | Moderately resistant | SBTIL 79 | xa13 + Xa21 | 1 | Resistant |
| SBTIL 24 | Xa21 | 3 | Moderately resistant | SBTIL 80 | xa13 + Xa21 | 1 | Resistant |
| SBTIL 25 | xa13 + Xa21 | 1 | Resistant | SBTIL 81 | xa13 + Xa21 | 1 | Resistant |
| SBTIL 26 | xa13 + Xa21 | 1 | Resistant | SBTIL 82 | xa13 + Xa21 | 1 | Resistant |
| SBTIL 27 | xa13 + Xa21 | 1 | Resistant | SBTIL 83 | xa13 | 3 | Moderately Resistant |
| SBTIL 28 | Xa21 | 3 | Moderately resistant | SBTIL 85 | xa13 + Xa21 | 1 | Resistant |
| SBTIL 29 | xa13 + Xa21 | 1 | Resistant | SBTIL 86 | xa13 | 3 | Moderately Resistant |
| SBTIL 30 | xa13 + Xa21 | 1 | Resistant | SBTIL 87 | xa13 | 3 | Moderately Resistant |
| SBTIL 31 | xa13 + Xa21 | 1 | Resistant | SBTIL 121 | xa13 + Xa21 | 1 | Resistant |
| SBTIL 33 | xa13 + Xa21 | 1 | Resistant | SBTIL 36 | xa13 + Xa21 | 1 | Resistant |
| SBTIL 34 | xa13 + Xa21 | 1 | Resistant | SBTIL 37 | xa13 + Xa21 | 1 | Resistant |
| SBTIL 35 | Xa21 | 3 | Moderately resistant |
3.4 Evaluation of pyramided lines for agro-morphological and grain quality trait
The BC2F2 generation was grown in the field and evaluated for various agro-morphological and grain quality traits. The pyramided lines were analysed to estimate the magnitude of the genetic variability for different morpho-physiological and grain and cooking quality. Significant variation was observed for all the traits viz. plant height, days to 50% flowering, days to maturity, effective tillers per plant, 1000 grain weight, panicle length, yield per plant, grain length, grain breadth, and L/B ratio. All the traits were at par with the recurrent parent. The introgressed line SBTIL80 showed maximum yield per plant of 67.3 grams with a maximum number of effective tillers, panicle length, and 1000 grain weight, which are considered to be yield-contributing traits. Similarly, introgressed lines SBTIL39, SBTIL25, SBTIL121, and SBTIL31 also showed yield per plant more than recurrent parent ranging from 42.4 g to 58.3 g. These observations allow us to conclude that agro-morphological traits play an important role in the plant's yield. The plant height of all the introgressed lines ranged from 130.9 cm to 137.5 cm and the minimum plant height was recorded in introgressed line SBTIL37. Most of the pyramided lines have 1000 grain weight higher than the recurrent parent (Table 3).
Agronomic performance of the introgressed lines along with parents.
| Genotypes | PH (cm) | DFF (No's) | DM (No's) | ET (No's) | PL (cm) | TGW (g) | YPP (g) |
| Ranbir Basmati | 145.7 | 89 | 108 | 18 | 31.4 | 22 | 21.6 |
| PAU148 | 121.4 | 106 | 141 | 21 | 33.2 | 25 | 30.4 |
| SBTIL21 | 133.9 | 93 | 135 | 14 | 34.1 | 21 | 31.9 |
| SBTIL25 | 133.1 | 91 | 129 | 25 | 33.3 | 22 | 47.4 |
| SBTIL27 | 137.5 | 91 | 132 | 16 | 35.7 | 21 | 23.8 |
| SBTIL29 | 134.7 | 94 | 132 | 07 | 28.3 | 22 | 15.2 |
| SBTIL30 | 132.4 | 89 | 134 | 20 | 31.2 | 26.5 | 32 |
| SBTIL31 | 134.3 | 92 | 121 | 19 | 35 | 29 | 42.4 |
| SBTIL33 | 135.3 | 98 | 114 | 18 | 32.2 | 19.5 | 15.4 |
| SBTIL34 | 134.6 | 84 | 118 | 18 | 32.4 | 20 | 22.4 |
| SBTIL36 | 134.7 | 101 | 132 | 07 | 31.6 | 19.5 | 19.2 |
| SBTIL37 | 130.9 | 85 | 119 | 09 | 34.1 | 21 | 22.5 |
| SBTIL38 | 135.3 | 93 | 122 | 10 | 39.9 | 20 | 24.4 |
| SBTIL39 | 131.1 | 93 | 136 | 19 | 32.4 | 22 | 58.3 |
| SBTIL42 | 132.0 | 89 | 135 | 09 | 32.9 | 23 | 27.3 |
| SBTIL49 | 134.1 | 85 | 121 | 07 | 34.9 | 20 | 14.7 |
| SBTIL80 | 132.7 | 100 | 132 | 21 | 33.0 | 26.5 | 67.3 |
| SBTIL81 | 135.6 | 99 | 137 | 13 | 33.5 | 23 | 24.4 |
| SBTIL82 | 134.8 | 99 | 137 | 11 | 34.9 | 24 | 27.3 |
| SBTIL85 | 134.5 | 107 | 143 | 14 | 31.9 | 24.5 | 12.3 |
| SBTIL121 | 132.2 | 90 | 120 | 17 | 35.0 | 23 | 43.7 |
| Sd(m) | 1.61 | 0.50 | 0.44 | 0.73 | 1.46 | 0.80 | 1.25 |
| CD | 4.78 | 1.48 | 1.26 | 2.17 | 2.39 | 3.73 | |
| CV | 1.70 | 0.76 | 0.49 | 7.15 | 6.24 | 5.03 | 5.99 |
3.5 Grain and cooking quality of introgressed line with respect to parents
The BC2F2 introgressed lines possessing all the target genes were also analysed for grain and cooking quality parameters. The KLBC for Ranbir Basmati and PAU148 was observed to be 7.13 mm and 8.16 mm respectively, while KBBC was 1.56 mm for Ranbir Basmati and 1.57 mm for PAU148. After cooking, KLAC values for Ranbir Basmati and PAU148 were 11.52 mm and 14.15 mm, respectively, while those for KBAC were 2.25 mm and 2.50 mm for Ranbir Basmati and PAU148, respectively. Interestingly, the KLAC and KBAC in introgressed line SBTIL121 were 13.98 mm and 2.48 mm, respectively, which is more than 2.46 mm in length and 0.23 mm in breadth than the recurrent parent (Fig. 7). The KLAC and KBAC were also measured in other introgressed lines that are depicted in Table 4.
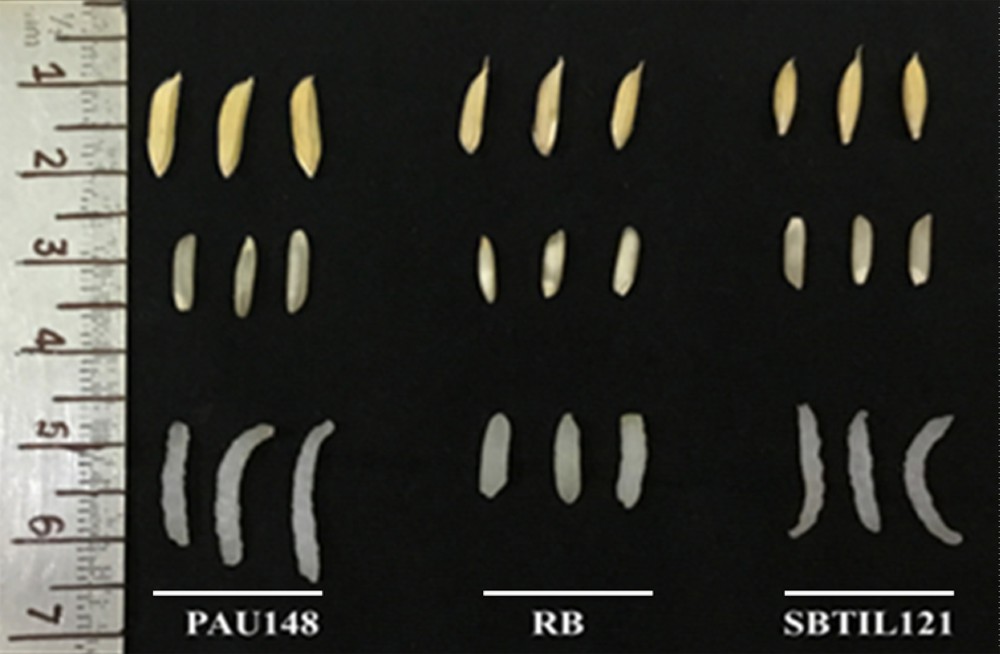
Comparison of grain and cooking quality of Ranbir Basmati (RB), PAU148 and SBTIL121. The SBTIL121 showed kernel length after cooking (KLAC) and kernel breadth after cooking higher than those of Ranbir Basmati.
Grain and cooking quality of the introgressed lines.
| Genotypes | GL (mm) | GB (mm) | L/B ratio | KLAC (mm) | KBAC (mm) | KLAC/KBAC ratio (mm) | Aroma |
| Ranbir Basmati | 7.13 | 1.56 | 4.56 | 11.52 | 2.25 | 5.12 | Strongly aromatic |
| PAU148 | 8.67 | 1.57 | 5.52 | 14.15 | 2.50 | 5.66 | Strongly aromatic |
| SBTIL21 | 7.13 | 1.74 | 4.09 | 13.18 | 2.65 | 4.97 | Strongly aromatic |
| SBTIL25 | 7.19 | 1.71 | 4.20 | 13.22 | 2.56 | 5.16 | Strongly aromatic |
| SBTIL27 | 7.51 | 1.52 | 4.92 | 13.49 | 2.15 | 6.27 | Strongly aromatic |
| SBTIL29 | 7.73 | 1.66 | 4.65 | 13.58 | 2.20 | 6.17 | Strongly aromatic |
| SBTIL30 | 7.42 | 1.62 | 4.56 | 13.32 | 2.25 | 5.92 | Strongly aromatic |
| SBTIL31 | 8.16 | 1.54 | 5.29 | 14.05 | 2.10 | 6.69 | Strongly aromatic |
| SBTIL33 | 7.73 | 1.61 | 4.80 | 13.59 | 2.15 | 6.32 | Strongly aromatic |
| SBTIL34 | 7.17 | 1.73 | 4.14 | 13.20 | 2.35 | 5.16 | Strongly aromatic |
| SBTIL36 | 7.48 | 1.69 | 4.41 | 13.44 | 2.36 | 5.69 | Strongly aromatic |
| SBTIL37 | 6.48 | 1.6 | 4.00 | 10.23 | 2.01 | 5.08 | Strongly aromatic |
| SBTIL38 | 7.37 | 1.67 | 4.40 | 13.36 | 2.24 | 5.96 | Strongly aromatic |
| SBTIL39 | 7.67 | 1.71 | 4.48 | 13.45 | 2.35 | 5.72 | Strongly aromatic |
| SBTIL42 | 6.68 | 1.62 | 4.12 | 10.25 | 2.34 | 4.38 | Strongly aromatic |
| SBTIL49 | 7.57 | 1.67 | 4.53 | 13.65 | 2.37 | 5.75 | Moderately aromatic |
| SBTIL80 | 7.63 | 1.72 | 4.43 | 13.68 | 2.45 | 5.58 | Strongly aromatic |
| SBTIL81 | 7.88 | 1.54 | 5.10 | 13.77 | 2.36 | 5.83 | Strongly aromatic |
| SBTIL82 | 6.85 | 1.53 | 4.46 | 10.26 | 2.35 | 4.36 | Strongly aromatic |
| SBTIL85 | 7.02 | 1.65 | 4.24 | 13.10 | 2.45 | 5.34 | Moderately aromatic |
| SBTIL121 | 7.97 | 1.64 | 4.25 | 13.98 | 2.48 | 5.63 | Moderately aromatic |
3.6 Genetic similarity in pyramided lines with the recurrent parent using SSR data
The cluster analysis of BC2F2 population was also done using molecular data, and the dendogram was formed using software NTSYS version 2.10. Cluster I consists of Ranbir Basmati and all the 19 pyramided lines that were sub grouped into cluster I-A, which contains Ranbir Basmati and SBTIL21, while cluster I-B is further divided into two sub-groups, cluster I-Ba and cluster II-Bb. Cluster I-Ba contains introgressed lines SBTIL25, SBTIL30, SBTIL29, SBTIL27, SBTIL31, SBTIL36, SBTIL37, SBTIL38, SBTIL39, SBTIL42, SBTIL49, SBTIL80, SBTIL81, SBTIL82, SBTIL121, and SBTIL85, while cluster I-Bb consists of SBTIL33 and SBTIL34. Cluster II contains PAU148, which is a donor parent (Fig. 8). The results revealed that all the introgressed lines are similar to those of Ranbir Basmati, a recurrent parent, and are grouped in same cluster.
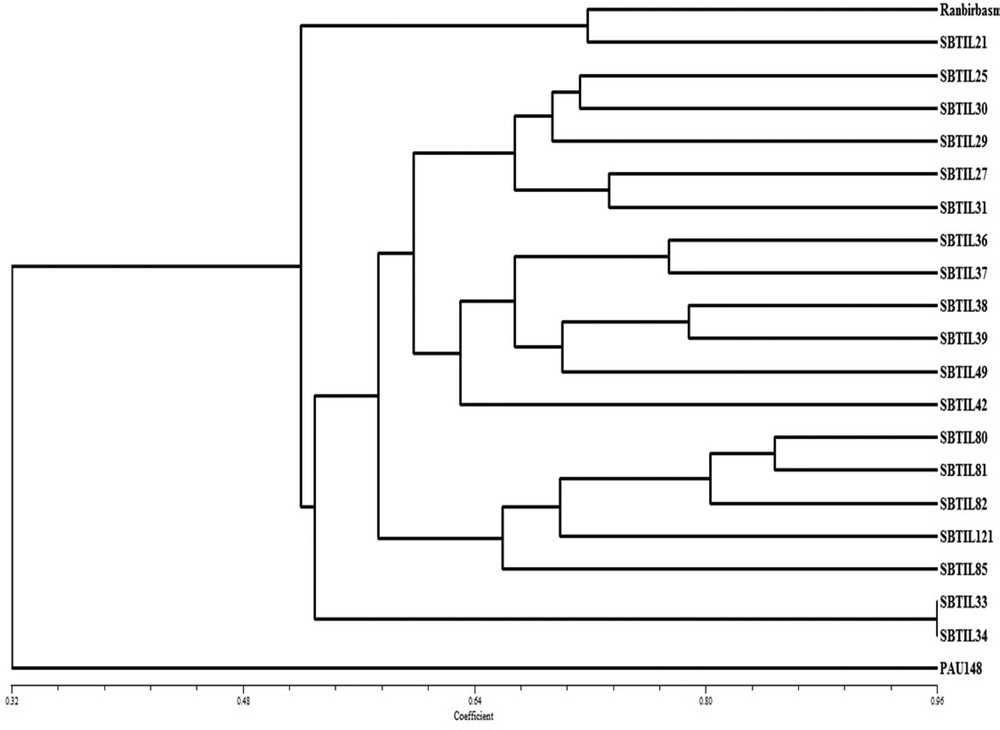
Dendogram illustrating the genetic relationship between pyramided lines of the BC2F2 population. The recipient parent (Ranbir Basmati) and other 19 introgressed lines are clustered into cluster I, whereas donor parent (PAU148) is in a separate cluster II.
4 Discussion
Marker-assisted backcross breeding is an efficient and precise system that allows for the selection of the recessive allele, selection at the seedling stage before a visible phenotype developed and pyramiding several useful traits in a single line without conducting traditional phenotypic evaluation. The most effective and environment friendly management strategy of combating these stresses is the exploitation of host plant resistance. MABB provides a great opportunity for precise transfer of desirable donor segment by minimizing the linkage drag into a recurrent parent. This strategy has been successfully utilized in several studies and, with the use of DNA markers tightly linked to the resistance genes, it is now possible to transfer beneficial alleles into the elite genetic background [24–26]. Combining MAS with phenotype selection for agronomic and grain-quality traits, BB resistance gene xa13 and Xa21 along with semi-dwarf sd1 gene was incorporated into the genetic background of Ranbir Basmati. The bacterial blight genes xa13 and Xa21 have been reported to provide resistance to multiple races of bacterial blight [10,27]. Therefore, these two resistance genes were selected and incorporated into the traditional variety Ranbir Basmati.
The semi-dwarf (sd1) gene was also successfully introduced in the genetic background of Ranbir Basmati along with BB resistance genes. In BC2F2 generation, a total of 19 genotypes were selected, carrying target genes (xa13, Xa21 and sd1), but apart from that, we observed that 41 genotypes were sd1 positive. These results were corroborated with the agro-morphological data, which shows that the genotypes carrying sd1 gene were short in stature than genotypes lacking sd1 gene. These results are in concordance with the findings of Rajpurohit et al. [12] and Luo et al. [28], who concluded that sd1 genes confer a semi-dwarf stature due to the loss of function of GA20ox-2 [3,4].
The improved lines of Ranbir Basmati in homozygous condition showed enhanced resistance under artificial inoculation of Xoo isolates. This study suggested that xa13 in combination with Xa21 was the most effective, with shorter lesion lengths than the single gene. However, Xa21 alone in some introgressed line showed resistance to BB. Sanchez et al. [14] and Gopalkrishnan et al. [29] suggested in their studies that the synergistic action or quantitative complementation between the resistance genes might result in increasing the level of resistance against the Xoo strain.
Marker-assisted background analysis of pyramided lines helped in identifying the line with maximum recovery of the recurrent parent genome. The highest genome was recovered in introgressed line SBTIL121 (86.9%) in BC2F2 generation. The graphical genotype of various BC2F2 progenies for background recovery showed that the genome recovery for various carrier chromosomes 1, 8 and 11 was more as compared to the non-carrier chromosomes. The chromosomes carrying target genes xa13 (chromosome 8), Xa21 (chromosome 11), and sd1 (chromosome 1) were given more emphasis as they have greater selection pressure for the donor parent allele at the target gene in each backcross generation. The percentage recovery of RPG in SBTIL121 was 86.9% in BC2F2 population, while the RPG percentage ranged from 70 to 86% in other introgressed lines calculated by GGT software. The low background recovery observed in some genotypes can be due to linkage drag, as explained by Baliyan et al. [30]. Also, the background selection using the SSR markers usually target the non-coding and heterochromatic regions and therefore may not quantify the recovery of the functional part of the genome [6]. However, the phenotypic selection, which indirectly targets the functionally expressed region of the genome, was augmented for hastening the process of reconstruction of the recurrent parent phenotype.
The agro-morphological data of 19 pyramided lines along with two parents revealed that the pyramided lines possess excellent features of recurrent parent with tolerance to bacterial blight disease. Analysis of variance for different morphological and grain quality characters revealed that all the introgressed lines along with the parents were at par with the recurrent parent and differ significantly for all the traits, viz. plant height, days to 50% flowering, days to maturity, effective tillers, panicle length, 1000 grain weight, yield per plant, grain length, grain breadth, and L/B ratio. It was interesting to note that, in our study, most of the pyramided lines were superior to the recurrent parent in terms of grain yield per plant, grain length, and grain breadth. Pradhan et al. [10] also reported that the yields of some pyramided lines were more than those of the recurrent parent, which may be due to the inheritance of some yield traits or QTLs from the donor parent transferred to the recurrent parent. The KLAC and KBAC were better for the recurrent parent obtained in BC2F2 generation. The trait KLAC and KLBC are under polygenic control governed by additive, dominance, and epistatic gene action showing the positive selection. Similarly, aroma, a key Basmati trait mainly governed by two recessive genes badh1and badh2, was strongly aromatic in all selected NIL populations. Combining marker-assisted foreground as well as background selection along with phenotypic selection was proved to be most effective breeding scheme, which has enormously increased the efficiency of breeding program.
The genetic similarities among pyramided lines and the parents were analysed by forming a dendogram. The dendogram formed using molecular data revealed that the 19 introgressed lines were closer to Ranbir Basmati, while the donor parent was clustered in another cluster. Similar results were reported by Rajpurohit et al. [12] and Sakthivel et al. [26]; in their study, they showed that NILs along with the recurrent parent are clustered together, while the donor parent makes a separate group.
The improved lines have desirable Basmati grain and cooking quality characteristics, in tandem with inbuilt resistance to BB and yield at par with the recurrent parent, i.e. Ranbir Basmati. These introgressed lines will be further backcrossed or evaluated under multi-location trials for release to farmers as improved Basmati cultivars. They will also be a unique source for BB resistance genes along with semi-dwarfing gene in future Basmati breeding programs. Overall, this study reports a successful introgression of BB resistance and semi-dwarf gene in Ranbir Basmati for resistance to BB with strong aroma. The phenotypic background selection implemented during MAS was effective in rapidly recovering the agronomic performance of the recurrent parent in selected lines. This shows that MAS accompanying phenotypic selection could be a reliable strategy in backcross breeding programmes.
5 Conclusion
In conclusion, this study identified 19 pyramided lines, among which an introgressed line ‘SBTIL121′, having all the target genes (xa13, Xa21 and sd1) combination, which is accompanied by high-level resistance to BB disease. Also, the introgressed line showed significant agro-morphological superiority in comparison to Ranbir Basmati. The study showed that the introgressed lines provide broad spectrum resistance to Xoostrains which can be further evaluated for release of a variety and they can also be used as BB and Semidwarf donor for introgressing in other elite basmati varieties.
Acknowledgements
The financial support provided under project by Department of Biotechnology, Govt. of India, New Delhi is highly acknowledged to carry out the research programme.


