1. Introduction
The Earth is undergoing rapid and unprecedented changes that surpass recorded historical events. Despite these transformations, the scientific community faces challenges in adequately reporting, documenting, and quantifying their wide-ranging consequences on biodiversity and the contributions of nature to people [1]. Pioneering monitoring programs have often targeted emblematic groups such as birds (e.g., American and British Bird Surveys) to represent the overall biodiversity, or focused on endangered habitats or species, as advocated by the Habitat or Bird directive of the European Union. While monitoring threatened species and understanding the long-term survival of bird species is crucial and has yielded significant discoveries, particularly in illustrating and quantifying time-lags and extinction debts [2, 3], these taxon-specific programs lack a holistic vision integrating biodiversity and ecosystem functions.
Biodiversity is not simply the sum of individual species but rather a complex interaction network involving various organisms, from soil bacteria to flying birds, influencing each other and collectively driving ecosystem responses to environmental changes [4, 5]. Detecting and attributing human-induced environmental modifications to biodiversity and ecosystems require adopting this holistic perspective [6]. However, constructing a monitoring program with a holistic vision is challenging, which explains the limited number of such initiatives (e.g., Lifeplan, NEON, eLTER, [7]).
First of all, to monitor biodiversity dynamics and attribute changes to potential pressures [8], a spatially-nested sampling scheme is essential. As such, elevational gradients in mountain environments provide a natural setting for tracking changes across different biodiversity compartments over time. Sharp changes in environmental conditions and human uses allow to track biodiversity dynamics along short distances. Under climate warming, what we will observe at a certain altitude in 50–70 years is likely what we nowadays observe 200 m below. However, these gradients must be replicated and distributed across multiple regions to best represent the diversity of geological/environmental/biogeographic contexts as well as the potential variability of biodiversity responses. Second, monitoring the whole biodiversity of ecosystems requires adapting the sampling protocols and tools to best capture the different compartments (below and above grounds) and their interlinkages. Soil sampling might require multiple samples in a given permanent plot, while a single acoustic recorder for the whole landscape is likely enough. Third, monitoring all biodiversity compartments across time requires another vision of re-sampling. Microbes or invertebrate responses to environmental changes are likely to happen at a different pace than forest trees or large herbivores. In other words, adopting for instance a strict 5-years interval sampling might not be the best strategy to capture very short dynamics of invertebrates. Fourth, it also requires combining and integrating different sampling strategies and tools to best represent the whole biodiversity. The complexity of soil biodiversity necessitates the use of high-throughput sequencing while other vegetation variables can be measured with more traditional field methods (i.e., floristic diversity, forest structure, and dead wood). Fifth, in situ environmental measurements are critical for attributing biodiversity changes, particularly in mountain environments where large-scale interpolations of climate and soil data are often inaccurate. Combining in situ measurements with high temporal and spatial resolution satellite imagery is thus required. Sixth, the quantification and statistical analyses of these data necessitate data integration tools and pipelines for effective use. Finally, visualization and validation tools are essential for engaging stakeholders and actively involving them in data collection and interpretation.
This paper presents an overview of the ORCHAMP program, a designed initiative for spatially and temporally explicit monitoring of biodiversity within the mountain ecosystems of the French Alps and Pyrenees. The primary objective of this program is to address the challenges mentioned earlier, focusing on the detection and attribution of biodiversity changes. To facilitate efficient data processing, archiving, and analyses for each biodiversity compartment and environmental data acquisition, we have developed specialized tools, pipelines and statistical analyses. Beyond this, ORCHAMP aims to enhance our understanding of biodiversity dynamics and structure, and to provide high-resolution predictions of biodiversity. Therefore, ORCHAMP federates a large number of institutional and academic partners in a framework of local collaborations with stakeholders and actors of wildlife conservation.
2. The elevational gradient as a primary observation element
ORCHAMP is conceptualized to systematically track the spatial and temporal dynamics of biodiversity. Given the backdrop of climate warming and land abandonment, we anticipate significant upward shifts in species distributions, leading to community reorganization and modified ecosystem functioning. Keeping track of these changes in mountainous environments entails monitoring the fluctuations in the spatial distribution and structure of biodiversity along elevational gradients. To address this, ORCHAMP strategically deploys permanent plots along multiple elevational gradients.
2.1. From permanent plots to an elevational gradient
The permanent plot represents the common unit of each sampling protocol, and within a plot, each protocol has its own location and extent (Figure 1). A permanent plot measures 30 × 30 m. An ORCHAMP elevational gradient is thus based on a certain number of permanent plots, spaced ∼200 m apart in altitude, with four to nine permanent plots per gradient. Ensuring the attribution of biodiversity changes is not compromised by uncontrolled sampling settings, elevational gradients are selected with relatively homogeneous slopes, minimal ruptures, changes of orientations, or strong discontinuity. Ideally, a linear transect from the bottom of the valley to the summit is sought, although achieving this is often challenging.
ORCHAMP monitoring scheme. Schematic illustration of an ORCHAMP permanent plot, measuring 30 × 30 m (adjusted for slope). The botanical survey is conducted along the central band (highlighted in yellow), incorporating a comprehensive survey spanning 1.5 m × 30 m, and using 2 × 150 pin-points for quantifying species abundances. Monitoring of trees and deadwood is implemented throughout the entire plot (depicted in orange). Acoustic and video recorders (shown in blue) are strategically positioned within or in close proximity to the permanent plots. Climatic hobos and TOMST are centrally located within the plot (star in the plot in dark pink), while soil sampling for eDNA and physico-chemical analyses (in brown) is performed 5 m down the botanical band (in yellow) and within three pre-selected sub-plots. A soil pit is excavated in the vicinity of the plot.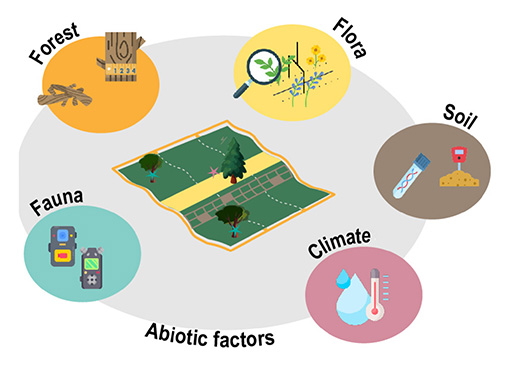
2.2. From an elevational gradient to multiple representative gradients of the region
Recognizing the limited statistical value of a single elevational gradient across a large mountain region, we selected gradients to be representative of the overall diversity of pedo-climatic conditions (Figure 2). This is achieved by collaborating with local partners to sequentially add new elevational gradients, ensuring they fill gaps in the multivariate environmental space of the French Alps and the Pyrenees (taken separately). The process began in 2016 with five gradients in the French Alps, and in subsequent years, with the addition of new gradients (Figure 3). In 2020, ORCHAMP initiated the re-survey of existing elevational gradients, slowing the addition of new ones in 2021, focusing on the complementarity of these gradients. Elevational gradients were also started in the Pyrenees in 2021, following the same sampling strategy as in the French Alps (Figure 3).
ORCHAMP spatial representation. Locations of the ORCHAMP elevation gradients through the French Alps and Pyrenees. Permanent plots are located 200 m of altitude apart of each other.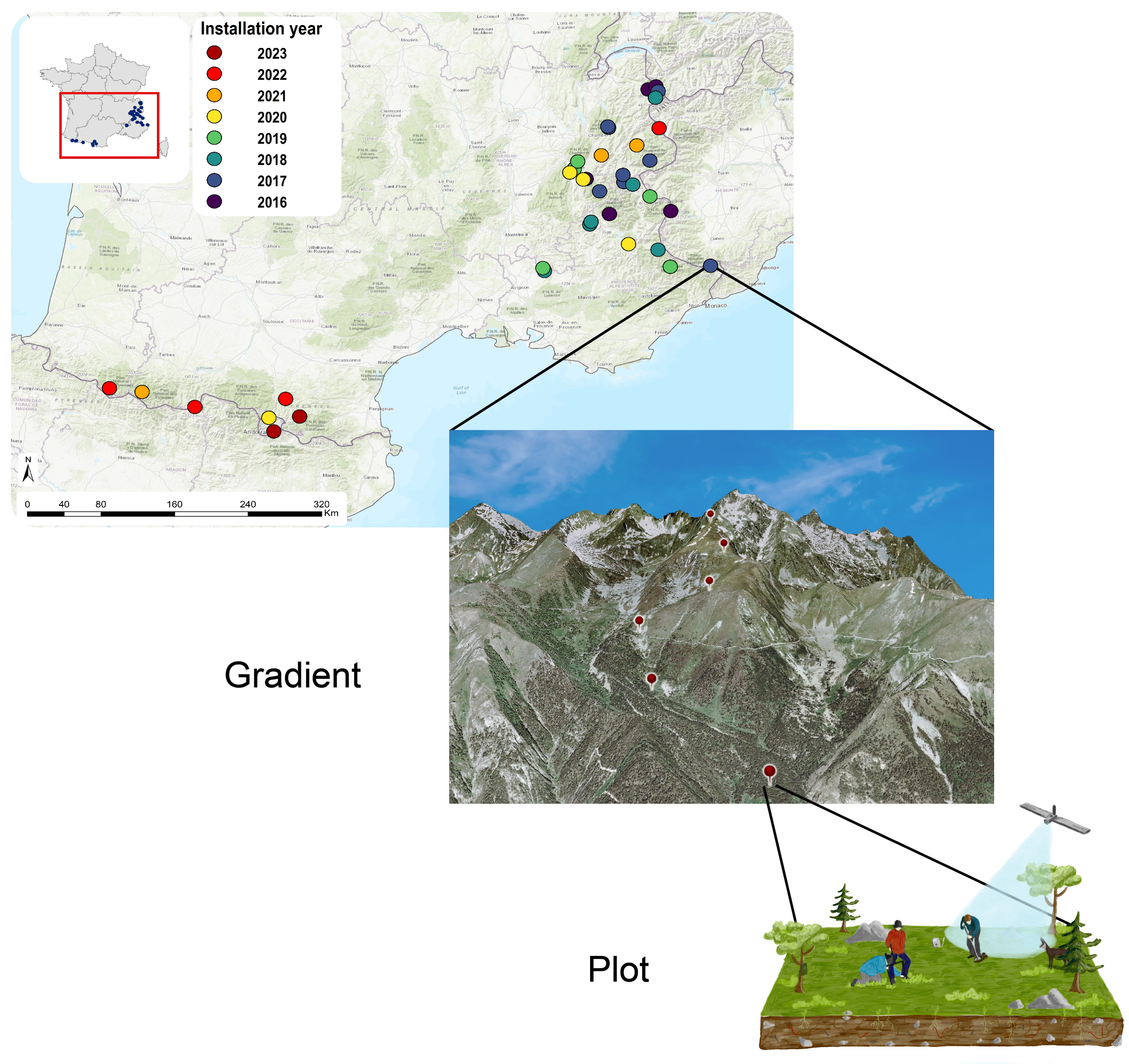
Number of ORCHAMP plots and gradients monitored. Cumulative representation of the elevation gradients and total number of plots across time, together with the number of plots sampled every year for the first visit or for a resurvey.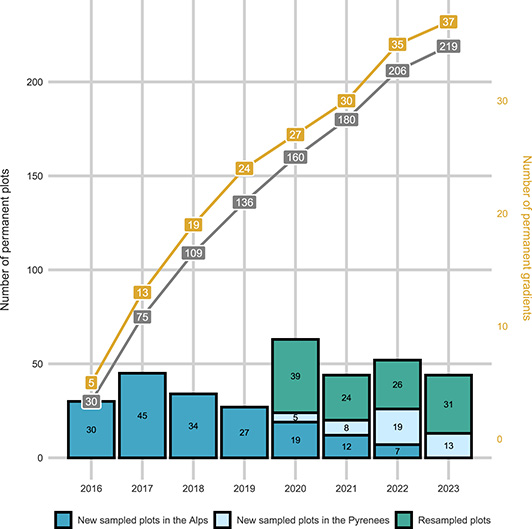
2.3. Resampling strategy
Given ORCHAMP’s diverse taxonomic targets, spanning bacteria to plants to large mammals, with varying life-history traits and responses to environmental changes, a rotative panel sampling approach was adopted. This strategy involves randomly selecting elevational gradients to be sampled each year, aiming for a relatively uniform time-interval across the gradients over the long run. The average time-interval is set at around 4–5 years. As ORCHAMP involves multiple academic and local partners, constraints are introduced in the random draw to ensure equitable distribution of workload, notably by limiting the number of gradients a specific botanical society can investigate per year. To enhance repeatability, a Shiny interface was developed to track the planned resampling strategy over the years.
3. Key monitored environmental features
To have the key environmental features to be contrasted to biodiversity, we follow relevant environmental features on each sampled plot. In a nutshell, we follow soil temperature, and more recently soil humidity, and soil physico-chemical properties for the in situ measurements. Once these data are combined with local climatic models and satellite images, they form our data-cube of environmental features (or essential environmental variables, [9], Figure 4).
Data integration and flow of information through ORCHAMP. Environmental features are collected through various sensors and techniques and form the environmental data cube. From below to above ground, environmental DNA, vegetation survey, acoustic and video recorders provide multi-taxa biodiversity data cube than can then be transformed into essential biodiversity variable data cube. Both environmental and biodiversity data cube are then used to detect and attribute biodiversity changes but also to provide predictions. Our dedicated soil food web ontology and data integration pipeline even allows constructing informative soil food webs.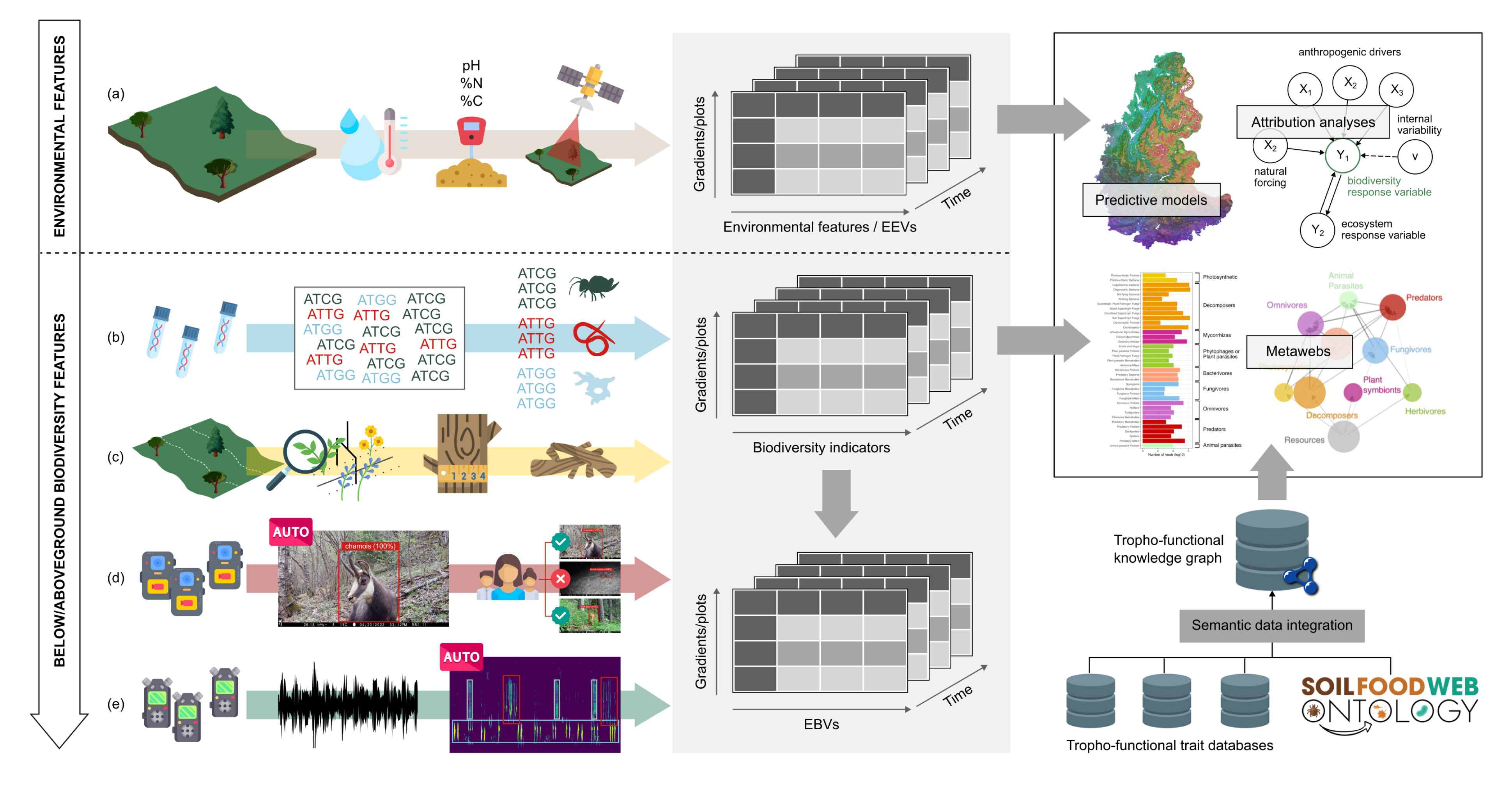
3.1. In situ climatic data and local climatic models
A better understanding of the local climate conditions experienced by living organisms is of paramount importance to anticipate biodiversity response to climate change [10, 11, 12]. The temperature measured within ecosystem compartments (i.e., understorey, soil) substantially differs from the air temperature measured by standardized weather station or modelled by regional and global climate re-analysis. The extent of this decoupling between microclimate and macroclimate exhibits large spatial variations in relation to land cover and topographical variations [13]. For example, densely forested stands play a more effective role in mitigating the diurnal and seasonal fluctuations of air temperature compared to open habitats, and this effect can explain contrasting responses of the understory plants to climate warming [12, 13].
In mountains, near-surface temperature variations are also largely determined by the duration of the snow cover period, which is mainly controlled by topographical variations and wind redistribution of snow [14, 15]. This leads to contrasting soil temperature regimes over short distances in above-forest ecosystems [14]. Understanding the patchiness of the thermal landscape in mountains is pivotal when investigating spatial and temporal changes in biodiversity and ecosystem processes.
All ORCHAMP sites have been equipped with miniaturized and standalone temperature data loggers (Hobo pendant UA, Onset Computer Corporation, Bourne, MA). Devices are buried at 5 cm below ground level to monitor the temperature of the topsoil layer. Soil temperatures are measured on an hourly basis and time series are updated every year since 2016 (Figure 1). Data have been organized according to FAIR principles (“Findability, Accessibility, Interoperability, and Reusability”, [16]) in the framework of a larger initiative regrouping all long-term monitoring sites in the French Alps1 . Since 2022, soil moisture data loggers have been added (TMS4, Tomst, [17]). As an example of ongoing analyses, Figure 5 compares soil temperature regimes for year 2021 and 2022 in 14 elevational gradients representing 67 ORCHAMP plots. In the South-Western Alps, the summer of 2022 stood out for the unprecedented duration of the heat wave [18]. In 2022, the combination of an early snow melt-out date (Figure 5a) and a warm summer (Figure 5b) led to a very high accumulation of growing days (Figure 5c) in all ORCHAMP sites. The results also show that forest vegetation has largely attenuated the impact of the 2022 heat wave on soil temperature (Figure 5).
Impact of the 2022 heat wave on the soil temperature regime of ORCHAMP sites. (a) Difference in snow melt-out date (SMOD) between 2021 and 2022. Most sites experienced an earlier SMOD in 2022. (b, c) Percentage rise in the average July temperature (b) and the Growing Degree Days (GDD) (c) in 2022 compared to 2021. GDD is the sum of average daily degrees above 0 °C. Green and gray symbols represent forested and non-forested vegetation, respectively. Results are displayed along the elevational gradient covered by 67 ORCHAMP sites.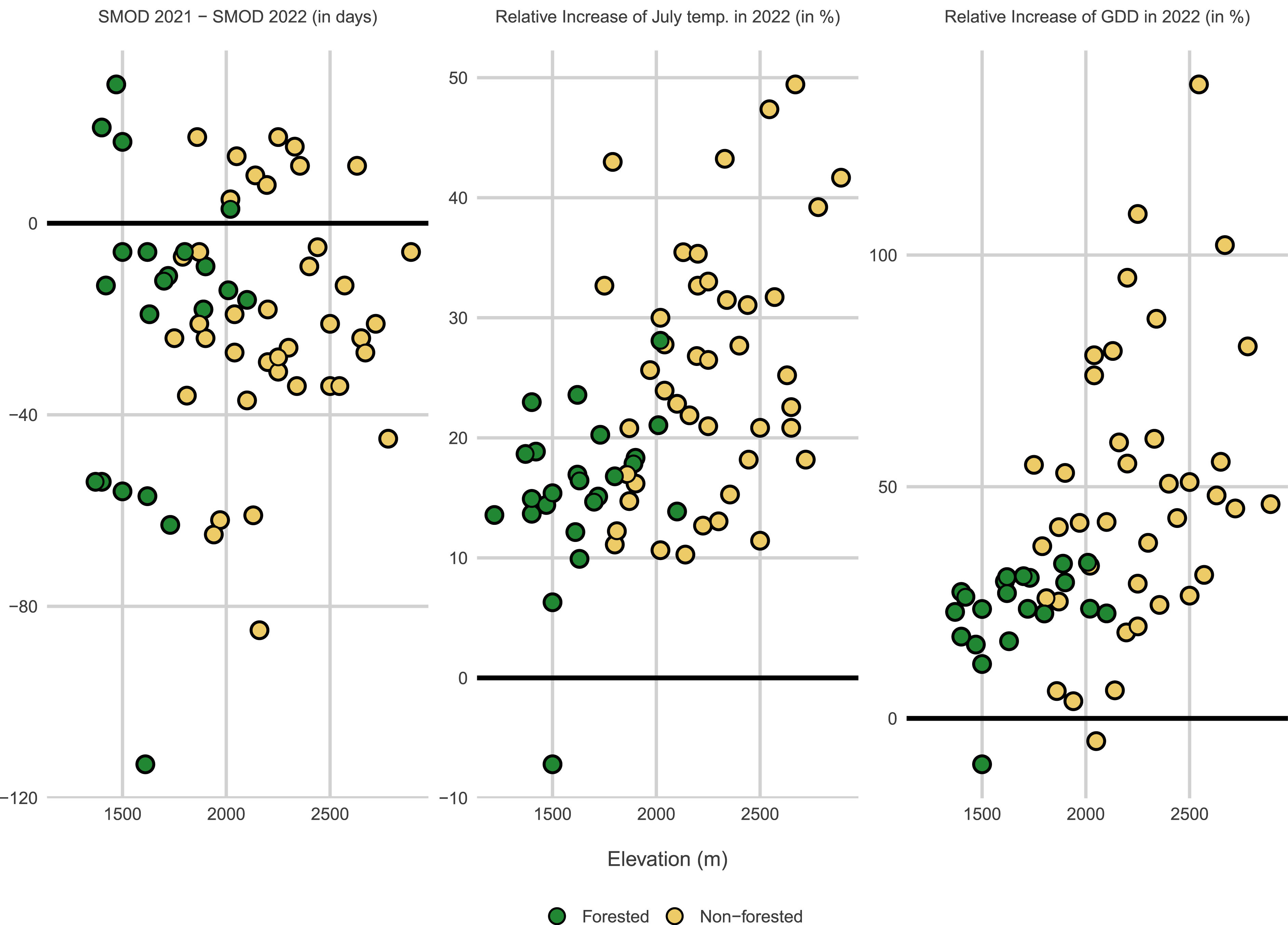
In addition to in situ measurements, climatic variables from the S2M re-analysis [19] are also extracted for each site. This dataset provides an hourly time series of meteorological data from 1958 to the present, including minimum, maximum, and mean temperatures for both soil and air, precipitations, solar radiations, and snow depth. Surface conditions are simulated as a function of elevation, slope, and aspect in the French Alps and Pyrenees.
3.2. Soil pit
On each permanent plot, a soil pit is dug (20 m downstream of the permanent plot to avoid disturbing the plot). Soils are described using the recommendations and coding procedures from the guideline for soil description [20]. Descriptions are carried out in the field using a specifically developed app (http://193.48.120.232:8080/descpedo/), which allows to standardize soil descriptions among operators and the descriptions to be directly integrated in the database. Soil samples are collected based on morphological horizons, emphasizing transitions, root profiles, and coarse elements. Where possible (and systematically for the 0–20 cm toposoil), known-volume soil samples, taken from cylinders, are added to reworked samples, allowing estimation of bulk density and stoniness for carbon stock calculation and water reserve assessment. Systematic sampling of soil parent materials is also realized, addressing both mono-parental (the large majority) and complex profiles with buried horizons and pedogenesis on various substrates. The samples are dried, sieved (2 mm), and quartered, with a portion stored in a sample library for future use (ORCHAMP pedological database).
Three main analyses are conducted on the pit soil samples:
- Conventional soil analyses (e.g., grain size, cation-exchange capacity, organic caron, total nitrogen and phosphor, pH) to define soil type and identify factors affecting biodiversity.
- Organic matter characterization (using infrared spectroscopy, Rock-Eval pyrolysis) to estimate organic carbon stock and stability.
- Mineral geochemistry analyses (quantitative X-ray fluorescence spectroscopy) to characterize the geochemical diversity of rocks, estimating the degree of weathering in soils.
Altogether these analyses provides comprehensive data on soil nature, leading to classification in soil reference systems [21]. Nearly twenty WRB soil references are covered, including variations in elevation, redox features, peaty soils, and more. The provided descriptions and pedological data are considered state variables influencing species distributions. For example, they make it very easy to position plots and transects within conventional gradients (acid rock–alkaline rock; soil with high or low water reserves, Figure 6). While the initial data aids in situating plots within conventional gradients, a second soil analysis campaign is planned for year n + 15 to assess medium-term changes, particularly in organic matter. This aims to observe statistically significant changes in organic matter stock and stability resulting from altered thermal regimes [22].
pH of the subsurface horizon in function of SiO2 bedrock content. Bedrock geochemistry provides a better estimate of soil properties than factorial information derived from geological maps.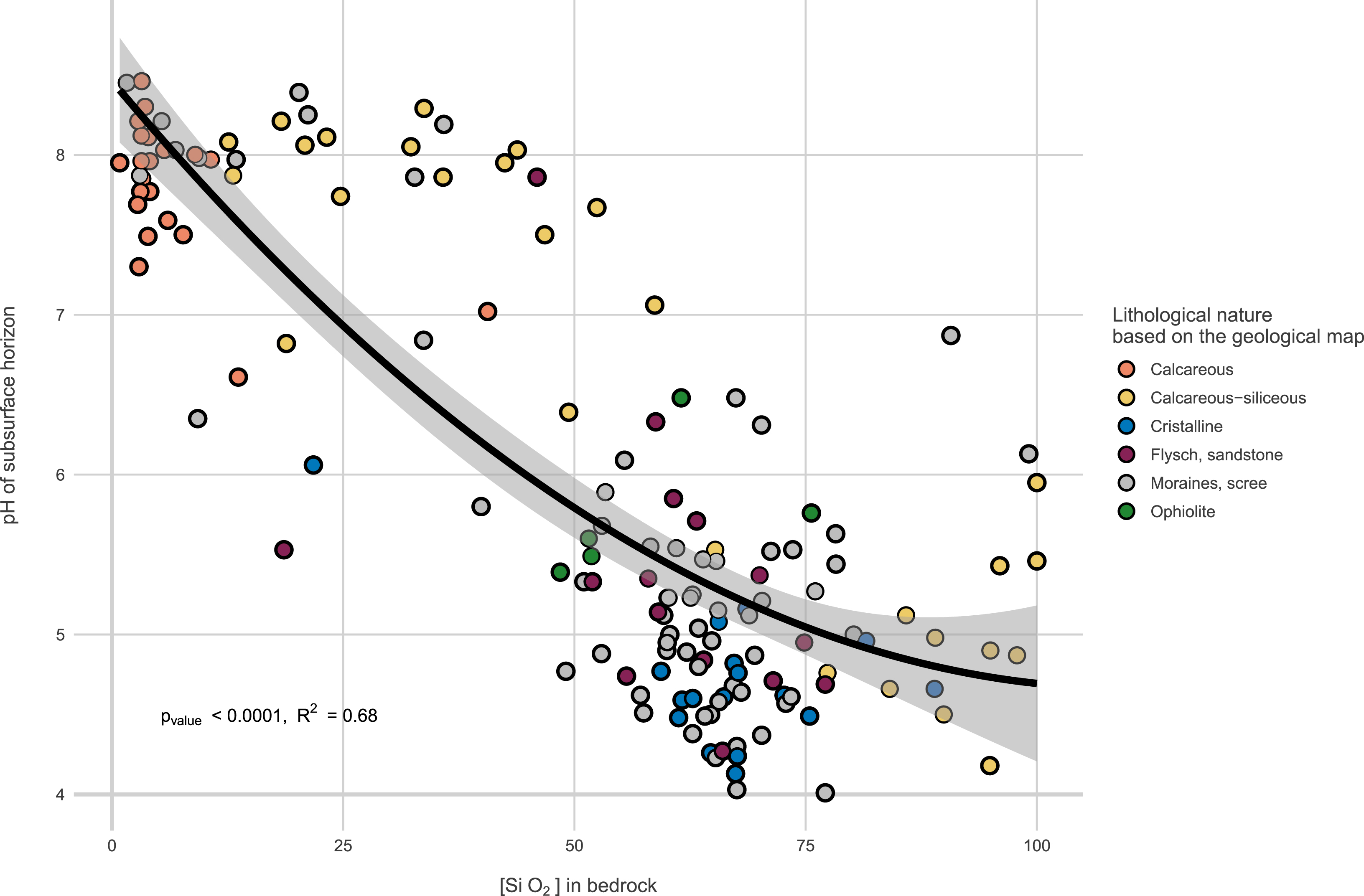
3.3. Soil physico-chemical properties
In each of the three 2 × 2 m soil subplots positioned 5 m below the inner transect of every sampling plot, ten soil cores are gathered and combined (Figure 1). A 15 g subsample is allocated for eDNA extraction. The remaining composite sample undergo a 2 mm sieving process and subsequent drying for the evaluation of physico-chemical properties. A section of the soil is finely ground to a particle size below 250 μm using an ultra-centrifugal grinder ZM 200 (ZM 200, Retsch). This facilitates the determination of total soil carbon and nitrogen contents through an elementary analyzer (Flash EA1112, Thermo Scientific). The remainder of the samples is dedicated to pH measurement in distilled water, adhering to the ISO 10390:2005 standard, utilizing a pH-meter (pH7110, inoLab). Soil organic matter (SOM) content is computed through Loss on Ignition, involving a 4-h incubation in a muffle furnace at 550 °C.
3.4. Remotely-sensed biophysical variables
Sentinel-2 has paved the way for cost-effective and high-resolution monitoring of vegetation, snow dynamics, and landscape configuration within ORCHAMP. This is achieved through a revisit time tailored to handle the prevalent heavy cloud cover at high altitudes and a ten-meter spatial resolution that aligns with the meso-topographic variability of our elevation gradients. Despite its advantages, the relatively short duration of the mission, commencing in 2017, poses limitations in tracking long-term trajectories essential for grounding biodiversity measurements in recent history.
The Landsat constellation offers the dual advantage of historical depth on a climatic timescale (more than 30 years of data) and spatial resolution tailored to mountain ecosystems. By combining these forces, we are able to derive high-resolution biophysical variables on all permanent plots and beyond, over the decades preceding the start of ORCHAMP. Employing annual maximum compositing of vegetation indices observations, such as Normalized Difference Vegetation Index (NDVI), we can generate multi-decadal, annual maximum NDVI (NDVImax) time series that act as indicators of vegetation changes. Positive trends in NDVImax, referred to as “greening”, signal an increase in vegetation greenness at the pixel scale, attributable to either an increase in vegetation biomass or productivity of preexisting vegetation [23] or shift in plant community composition [24, 25]. In mountainous regions, remote sensing is a favored tool due to the large study areas involved and the obvious difficulty of access and is therefore increasingly employed to observe how these ecosystems respond to ongoing climatic and usage changes [24, 26, 27]. Notably, Choler et al. [26] demonstrated the spatial variability of greening trends in above-forest habitats using MODIS satellites, with evidence that greening is widespread at the scale of the European Alps, but that high magnitude greening is mostly occurring in north-facing and sparsely vegetated slopes between 1900 and 2400 m, a phenomenon further corroborated by subsequent studies [27]. As an example of ongoing research, Figure 7 displays the NDVImax trends observed from 1984 to 2023 along the elevation gradient across all ORCHAMP sites obtained using all Landsat 5 TM, 7 ETM+ and 8 OLI images available in Google Earth Engine. We obtained the annual maximum NDVI and computed trends using a Theil-Sen estimator (R package mblm). Our results reveal that forests above 1800 m are positively responding to air temperature increases over the past 40 years, whereas forests at lower elevations are either not benefiting or adversely affected, aligning with recent studies [28]. Across all above forest sites (>2100 m), a positive vegetation response is observed, with almost no negative trends, consistent with the ecophysiology of alpine ecosystems amid warming [29] and supported by recent observations [25, 27].
NDVImax trends from 1984 to 2023 along the elevation for all ORCHAMP sites in the French Alps. Green and yellow points correspond to forested and non-forested sites, respectively. Curves are obtained from Generalized Additive Models (GAM) with shaded areas corresponding to standard error estimates. Green curve is only for forested sites while black curve is for both forested and non-forested sites. The color bar to the right of the panel is simply a qualitative indicator of the degree of browning or greening.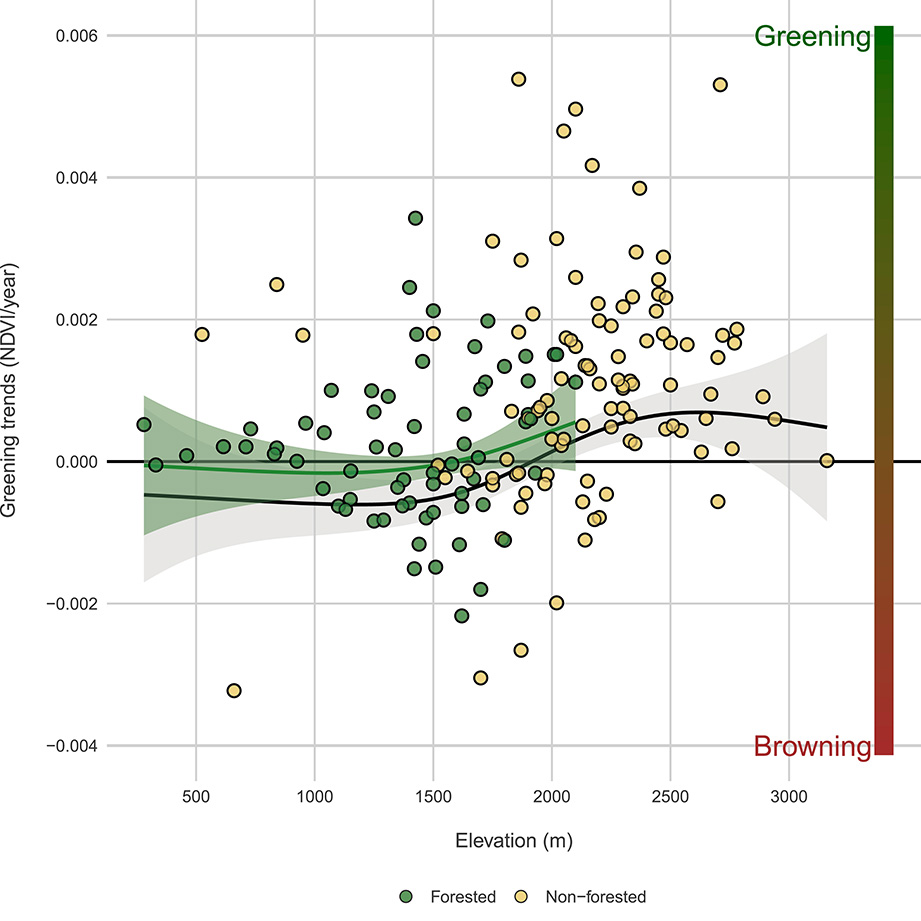
4. Key monitored biodiversity features
ORCHAMP mixes various biodiversity measurements from traditional botanical surveys to camera-traps with automatic assignments. The overall idea is to monitor biodiversity from the below-ground compartments through environmental DNA metabarcoding targeting the whole soil biodiversity, vegetation through botanical survey and remote sensing, to the above-ground compartment through acoustic and camera-traps sensor targeting mammals and birds. Several protocols have been defined for this purpose, some of which are mandatory and implemented on each transect, while others are optional (see Appendix A for the full list of variables monitored).
4.1. Plant vegetation survey and forest monitoring
4.1.1. Vegetation survey
On each plot, a presence–absence survey is conducted along the central band (Figure 1) to characterize the dynamics of plant species assemblages in time, including rare species. Although this protocol targets species presence–absence, the overall area (900 m2) allows the detection of rare species. So far, the lowest number of species richness was six (a dense spruce forest) and the largest was 93 (rich subalpine grasslands).
4.1.2. Pin-point transects
To monitor relative species’ abundance, a pin-point sampling is also carried out along the central band. At every 20 cm increment, all plant individuals in contact with the pins placed at the two measurement points (one 25 cm upslope and the other 25 cm downslope from the transect) are identified and recorded. This process yields a total of 300 pin-points per plot, each potentially involving multiple contacts with individual plants. So far, the lowest number of recorded individuals was two (a high summit at 3000 m) and the largest was 1472 (rich subalpine grasslands). This protocol is an adaptation of the Gloria “Downslope plant survey” [30]. In theory, this heavy protocol rather targets the dominant species and allows following their relative abundance trough space and time, but moderately rare species can also be detected.
4.1.3. Forest monitoring
The structure of both alive and dead trees is a crucial element of biodiversity and influences all other biodiversity components monitored in ORCHAMP [31]. The forest structure monitoring protocol employed is tailored from the one utilized in the French forest reserves monitoring network [32], ensuring inter-comparability.
Live trees: On each plot, trees are identified and located, and their Diameter at Breast Height (DBH, 1.30 m from the ground). Individuals with a diameter larger than 30 cm are surveyed in the entire 30 × 30 m plot, those with a diameter larger than 7.5 cm are only surveyed in the area 5 m upwards and downwards of the central band. Trees smaller than 7.5 cm with a height >1.3 m) are surveyed on the central band only.
Deadwood: Three deadwood types are documented: fallen logs, standing snags (height ⩾ 1.30 m), and stumps (height ⩽ 1.30 m). Each deadwood piece is identified to species whenever possible, and categorized based on (i) wood decay level (hard, less than 50% rotten, more than 50% rotten) determined by a knife penetration test [33]; and (ii) bark presence (intact, more than 50% attached bark, less than 50% attached bark, no bark). All deadwood (standing and fallen) with a minimum diameter of 30 cm is measured in the 30 × 30 m plot. In the 5 m zone above and below the central line, stumps and snags with a diameter between 7.5 cm and 30 cm are measured (mid-height for stumps, 1.30 m height for snags). Lastly, along the central line (see Figure 1), logs intersecting the line and with a diameter between 7.5 cm and 30 cm are measured using the Line intersect sampling method.
Tree-related microhabitats: A recent addition to the forest protocol concerns Tree-related Microhabitats [34], that are tree-borne singularities that provide food, shelter and nesting grounds for a variety of species (Figure 8). We inventory tree-related microhabitats on all living and standing dead trees with a DBH > 7.5 cm (except stumps) following the typology of Larrieu et al. [34] at its finest scale (47 types). We also quantify the abundance per tree of each microhabitat type.
Changes in epiphytes along elevation. Green, blue and red lines represent bryophytes, lichens and ivy and other lianas, respectively.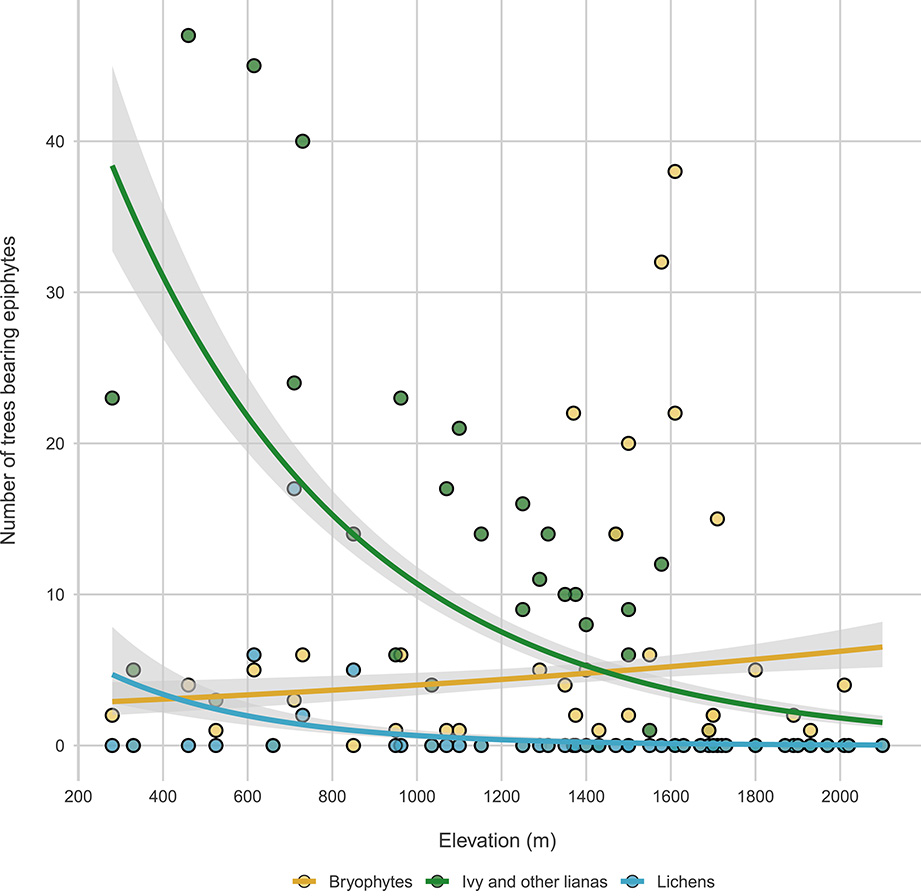
4.2. Below-ground biodiversity from environmental DNA
For each of the three subplots, 15 g aliquot are sub-sampled from the composite soil samples extracted with the drill (Figure 1) and then dried using silica gel. After each annual sampling campaign, eDNA extractions are performed and analyzed at the Laboratoire d’Ecologie Alpine following a standardized protocol [35]. Six DNA markers are employed to characterize the whole soil biodiversity, including two universal markers (euka02 for eukaryote, bact01 for bacteria) and four clade-specific markers (fung02 for Fungi, inse01 for Insects, olig01 for Oligochaeta, and coll02 for Collembola, [36]). DNA extraction, sequencing and all bioinformatic pipeline follow well-established and standardized pipelines (Supplementary Materials, [37]).
Environmental DNA metabarcoding offers cost-effective means to obtain extensive data on the whole soil biota. This big data is characterized by its heterogeneous taxonomic resolution, which is directly related to the discriminatory power of the DNA markers used [38]. To provide a more concise representation that still provides insights into the local structure and composition of soil communities, we assign the retrieved taxonomically annotated MOTUs to trophic groups of organisms that feed on the same food sources and have the same consumers [39, 40]. We subsequently link these groups based on known interactions to construct food webs that consider multiple trophic groups (nodes) and their linkages across trophic levels (links) [37].
The construction of food webs from large eDNA datasets was so far hampered by the lack of an overarching framework for classifying all soil biota based on their feeding preferences, and the scarcity and dispersion of trophic information for soil organisms in a multitude of data sources. To address the first challenge, we first developed the Soil Food Web Ontology (SFWO) to establish consensual and formal definitions for the range of concepts relevant to soil food webs [41]. The SFWO provides logical (in the sense of mathematical logic) definitions for more than 160 trophic groups across different taxonomic groups. This allows using automated reasoning to classify soil-associated consumers into trophic groups in consistent way. In parallel, and to address the second challenge, we developed an ontology-based data integration pipeline to facilitate the construction of biodiversity knowledge graphs from multiple data sources [42]. This pipeline was used to build a knowledge graph integrating information about the trophic interactions and feeding-habits of soil organisms from a dozen data sources. This graph provides a unified access to multisource trophic information across taxonomic groups and trophic levels. Within the knowledge graph, the integrated data are “semantified” (i.e., linked to concepts in the SFWO). This solves the problem of the semantic heterogeneity between data sources and allows the use of automated reasoning to deduce logical consequences (e.g., belonging to a trophic group) from explicitly-stated facts about trophic interactions and/or diets.
This knowledge-based approach greatly facilitates the reconstruction of informative metawebs, aka potential networks containing all the detected trophic groups and their potential interactions, from community composition data [37] and to analyze their changes in functions of environmental factors or habitats (Figure 9), or through time.
Soil food web variation along ORCHAMP gradients. Example of soil food web reconstructed through our automated pipeline and the relative frequency of the different trophic groups. Colors correspond to the trophic class to which the trophic groups belong to. The size of the circle corresponds to the relative frequency compared to the maximum ever observed through the whole set of the plots. Ew: earthworms, B: bacteria, F: fungi, N: nematods, P: protists. P-bact: protist bacterivores, F-ecm: ectomycorrhizal fungi etc.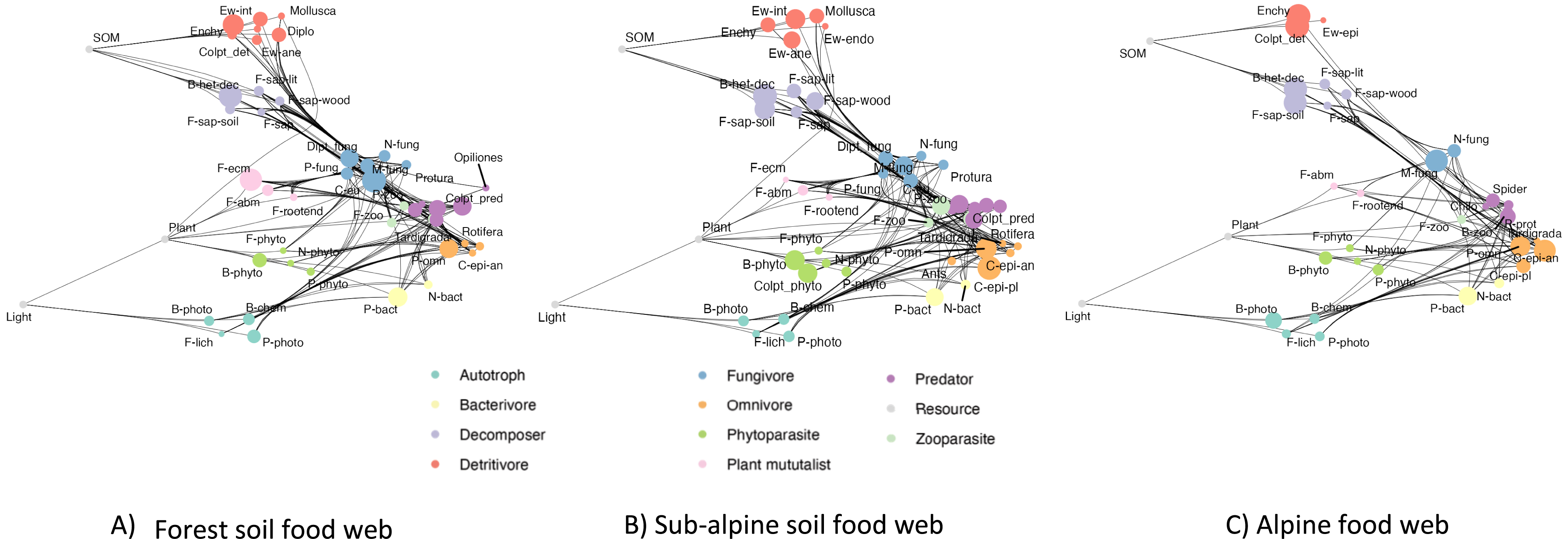
4.3. Above-ground biodiversity—from camera-traps to automatic recognition, validation and diffusion
At ORCHAMP, our focus revolves around the observation of large mammals during the summer as they pass through or closely inhabit the permanent plot. To achieve this, we employ SPEC Elite HP4 (Browning) cameras strategically positioned on trees within forest plots and on rocks or wooden poles in open plots, ideally situated approximately 60 cm above the ground. Two camera-traps are installed on each permanent plot, oriented preferably towards animal tracks whenever possible.
This monitoring protocol underwent testing in 2021 across 11 permanent plots and was officially initiated in 2022. Throughout 2022 and 2023, we extended our surveillance to 69 plots spanning 14 gradients. The camera traps are programmed to capture sequences of three images for each detection event, enhancing the accuracy of species identification.
Leveraging recent advancements in AI technology, we employ a two-step process, each step relying on a deep learning model. In the first step, using the MegaDetector (version v5.0a; https://github.com/agentmorris/MegaDetector/releases/tag/v5.0), we detect animals of any species, as well as humans and vehicles, sorting out empty images. Images with the highest confidence score above a threshold of 0.5 are retained, forming a bounding box that contains the most crucial element for classifying the entire image. In the second step, when the detected element is an animal, we use the DeepFaune classifier [43], a deep-learning-based species classification model (version v1.0.0) specifically designed for European fauna. Notably, this model was trained using images from various partners, including ORCHAMP, ensuring excellent accuracy on our images. Finally, the DeepFaune add-on dedicated to image sequence management [44] is employed for a single species prediction for each sequence.
The entire pipeline for camera-trap data is designed to be end-to-end. Images are transferred from SD cards to the Grenoble University computing center, where they are dynamically processed using the presented AI-based approach. Metadata, including classification results (empty/human/vehicle or any class in DeepFaune), are stored and can be inspected using a web interface developed with Kibana. Additionally, to enhance the confidence in image classification, an additional validation module inspired by citizen science initiatives like Zooniverse (e.g., project Wild Mont-Blanc) has been integrated. ORCHAMP staff can use a software app to review sequences with low-confidence AI predictions (DeepFaune score below 0.8, representing less than 20% of non-empty images). This allows staff to vote for or against proposed predictions and make corrections if necessary, ensuring a high level of confidence in image classification through a combination of AI and manual verification.
4.4. From passive acoustic monitoring to sonotype diversity and soundscapes
Passive Acoustic Monitoring (PAM) is an emerging sampling method that consists in collecting sound in the field with autonomous recording units [45, 46]. This method allows for a non-invasive survey with high spatio-temporal replication [47] and can target all soniferous species (e.g., birds, mammals, anurans, arthropods, fish) but also environmental noises and physical processes (e.g., rain, wind or anthropogenic noise) in an environment [48].
Within ORCHAMP, we focus on audible terrestrial animals and sounds present in or close to the plots. We record 1 min every 15 min, every other day for three-four months over summer with SMmini (Wildlife Acoustics Inc, Concord, MA, USA). The recorders are set at 48 kHz sampling rate, which allows detecting sounds up to 24 kHz. We install the recorders at 1.5 m from the ground on trees in forested plots or on wooden poles in plots located in open habitats. This protocol was tested in 2019 and initiated in 2022. Over 2022 and 2023, 52 plots over ten gradients were recorded. The recordings are stored on the University of Grenoble Alpes server and a pipeline automatically calculates a set of acoustic indices, which does not require any species annotation. The result of these analyses is stored as metadata for each recording.
Studying the structure and dynamics of acoustic communities using passive acoustic monitoring requires the ability to determine the composition of the community from audio recordings. The amount of raw audio data collected as part of ORCHAMP is growing rapidly, making expert-based acoustic animal identification impractical and motivating the search for automated approaches. Most existing techniques for identifying animal calls from ecological soundscapes perform a species-by-species analysis (e.g., BirdNet, [49]), with recent studies targeting birds, anurans, mammals, and insects [47, 50, 51]. The simultaneous detection of animal vocalizations from multiple taxonomic groups remains a challenge, especially in the absence of the labeled data essential for training the state-of-the-art species identification models relying on supervised learning. A promising alternative is the use of unsupervised learning to identify acoustic OTUs from audio recordings [52]. In this approach, an acoustic event detector extracts regions of interest from noise-reduced spectrograms. Spectral and cepstral features are then extracted and clustered, with the different clusters being (hopefully) associated with different species calls. We are currently developing our own version of this unsupervised pipeline, combining deep neural network-based acoustic event detection and feature extraction to overcome some of the weaknesses of the original implementation.
5. Data integration and data cubes
Each raw data and associated metadata collected in ORCHAMP are collected and stored using an online web application developed at LECA and stored in the ORCHAMP relational database, except for large and specific data sets (eDNA, acoustic, camera) where data are stored on independent storage platforms owned by Univ. Grenoble Alpes. For eDNA a specific meta-database is also available to store laboratory metadata.
For common indices and summarized data calculated from raw data, scripts are available for the ORCHAMP Consortium through an institutional Gitlab repository.
Raw data are made available for each type of data and protocol and for each plot using an R-Shiny application (interactive web applications straight from R, https://orchamp.osug.fr/api/general), where user can select the data needed and download .csv files. Graphs are also automatically generated to visualize the results.
All collected data through time form data cubes of Essential Variables, which are sparse given the fact we do not resample every year all plots and that re-sampled plots are drawn at random (Figure 4). Yet, those data cubes, which contains both biological and environmental data are extremely suited to build statistical models to understand the main drivers of biodiversity [39, 53], to understand biotic interactions [54] and their effects on ecosystem processes [53] but also to predict them through space and time. Last but not the least, these data can be used to detect and attribute biodiversity changes through causal models [8].
6. ORCHAMP—a playground for additional projects
ORCHAMP represents a real-world laboratory which also may serve as an expansive playground where researchers can develop supplementary projects, within the framework of ORCHAMP primary objectives and with the benefit of shared datasets of the consortium. Such side projects represent key advantages for assessing complexity and diversity of mountain ecosystems facing global change, and offer a fertile ground for innovative approaches beyond the common ORCHAMP design. ORCHAMP encourages collaborative and interdisciplinary research methodologies, where ideas from various fields can converge to study multifaceted questionings.
Hereafter, we present some additional side-projects within ORCHAMP.
6.1. Traditional monitoring of soil macroinvertebrates
Soil macroinvertebrates, whether in the topsoil or at the soil surface, serve as crucial connectors between soil food webs (investigated through eDNA analysis) and biodiversity in the above-ground environment (monitored using acoustic or camera traps). The macrofauna plays a pivotal role as regulators in both the brown and green pathways of energy flows in terrestrial ecosystems and represents significant food sources for various vertebrates, encompassing diverse taxonomic groups.
Sampling of soil macroinvertebrate communities occurs in early autumn through two standardized methods. Firstly, macroinvertebrates are manually sorted from four soil monoliths (25 × 25 cm), reaching depths of up to 20 cm when feasible. During the same period, macroinvertebrates at the soil surface are collected using six pitfall traps. Taxonomic classification is carried out at the highest resolution possible. Earthworms and ground beetles, representing distinct trophic groups, are individually weighed based on a trait directly linked to fitness.
Leveraging these data and the data cubes from ORCHAMP, comprehensive analyses will reveal the key drivers influencing multi-trophic community composition changes. It will allow disentangling bottom-up (resource-driven) and top-down (regulation by climate, vertebrate predators, etc.) factors at nested spatial scales, from local to regional scales. Additionally, it provides a platform to scrutinize the distinct roles played by intraspecific and interspecific trait variability in explicating the observed patterns. Furthermore, changes in intraspecific trait variability will serve as early signals of population adaptive responses to environmental changes. More specifically, we will assess the complementarity of sampling methods (environmental DNA and hand-sorting) for characterizing the response of earthworms to environmental gradients, providing elements for implementing future large scale monitoring schemes.
6.2. Retrospective approaches
ORCHAMP has also enabled the emergence of retrospective approaches to biodiversity, using paleoecological and historical ecology disciplines along selected transects. For instance, the study of anthracological fossil contents in soil pits (across 4 transects) will allow the assessment of the past presence (sometimes on a multi-millennial scale) of structuring forest taxa in the vegetation cover. Similarly, multi-proxy analysis of several Holocene lake sediments (sampled near the transect), aims to trace past dynamics of mountain ecosystems, changes in land-use, erosion and past climate (across various temporal scales), focusing on the age, the intensity, and the recurrence of high-altitude zone openings under the combined influence of past land-use and Holocene climatic variability.
7. Conclusions
Started in 2016, we expect ORCHAMP to become a cornerstone for the biodiversity monitoring in mountain environments. We are currently in discussion to set up new elevational gradients in Corsica mountains and a further logical expansion will involve the whole European Alps. Such an endeavor will require building long-term international collaborations with scientists and stakeholders from other countries. Yet, while we have been able to foster collaborations between national parks, reserves, local actors and researchers, building such a consortium across several countries will require more stabilized fundings and regulatory processes.
Declaration of interests
The authors do not work for, advise, own shares in, or receive funds from any organization that could benefit from this article, and have declared no affiliations other than their research organizations.
Acknowledgments
The ORCHAMP project is only possible thanks to the many people that participate in the field work and in the lab procedures, including students, interns, postdoctoral fellows, field assistants, park managers and rangers. The complete list of funding is display in Appendix B, but we would like to acknowledge support from the French Agence Nationale de la Recherche (ANR) through the GlobNet, TransAlps & EcoNet projects (ANR-16-CE02-0009, ANR-18-CE02-0010-01), from two “Investissement d’Avenir” grant (Idex UGA: ANR-15-IDEX-02; Montane: OSUG@2020: ANR-10-LAB-56, MIAI@Grenoble Alpes: ANR-19-P3IA-0003), Grenoble Alpes Métropole, the Agence de l’eau, the Isere departement and the Office Français de la Biodiversité (OFB). Support was also received from the Lautaret Garden-UAR 3370 (Univ. Grenoble Alpes, CNRS, Jardin du Lautaret, 38000 Grenoble, France), member of AnaEE-France (ANR-11-INBS-0001AnaEE-Services, Investissements d’Avenir frame) and of the LTSER Zone Atelier Alpes, a member of the eLTER-Europe network.
Appendix A
See Table A.1.
List of the different measures collected within ORCHAMP, their frequency, whether there are optional or not, to which Essential Climate or Biodiversity variables, ECV and EBV, respectively, they belong to
| Compartment | Corresponding protocol | Survey/method type | Measured parameters | Area | Frequency | Additional information | Essential variables (EBV/ECV) |
|---|---|---|---|---|---|---|---|
| Climat | In situ climatic data | Stand alone logger | Soil temperature and soil moisture measured at 5 cm below ground | 1 logger per plot | Hourly | ECV: Land/Biosphere/Soil moisture & Land surface temperature | |
| Climat | Local climatic models | S2M meteorological and snow cover reanalysis | Temperature (min/max/mean) for soil and air, precipitations, solar radiations, snow depth, evapotranspiration … | At the plot level (from 1958) | Hourly | ||
| Environment | Pit soil | Cylinder sampling | Estimation of bulk density and stoniness | 1 “pit” subplot outside each plot | Once per plot | ||
| Environment | Pit soil | Soil physico-chemical properties | Grain size, Cation exchange capacity, Corg, total N, P, pH … | 1 “pit” subplot outside each plot | Once per plot | ||
| Environment | Pit soil | Soil organic carbon properties | Thermograms obtained using Rock-Eval® thermal analysis | 1 “pit” subplot outside each plot | Once per plot | ECV: Land/Biosphere/Soil carbon | |
| Environment | Pit soil | Mineral geochemistry analyses | Elemental analysis using quantitative X-ray fluorescence spectroscopy | 1 “pit” subplot outside each plot | Once per plot | ||
| Environment | Top soil | Soil physico-chemical properties | Soil pH, soil organic matter, total soil carbon, nitrogen and phosphorus contents | 3 “topsoil” subplot for 2 ∗ 2 m per plot | Each resampling year | ||
| Environment | Top soil | Soil biological activity | Potential extracellular enzymatic activities | 3 “topsoil” subplot for each 2 ∗ 2 m per plot | Each resampling year | Optional | |
| Environment | Remote sensing | Satellite images from Sentinel-2, Landsat and MODIS | Multi-spectral bands | At the plot, gradient and regional level | Each resampling year | EBV: Terrestrial/Ecosystem function/Primary productivity & Ecosystem phenology | |
| Environment | Remote sensing | Satellite images from Sentinel-2 & Landsat | Multi-spectral bands | At the plot, gradient and regional level | Each resampling year | EBV: Terrestrial/Community composition/Trait diversity | |
| Topography | Plot description | Topographical measurements | Altitude, exposition, slope | At the plot level | |||
| Biodiversity | Plant | Presence–absence | Species name (Taxref V.14) | 3 “botany” subplot of 3 ∗ 10 m per plot | Each resampling year | EBV: Terrestrial/Species populations/Species distributions | |
| Biodiversity | Plant | Pin-point transects | Species name for each contact (Taxref V.14) | 300 pinpoints per plot (within “botany” subplot) | Each resampling year | EBV: Terrestrial/Community composition/Community abundance | |
| Biodiversity | Forest | Live tree inventories | Species name, location in the plot, Diameter at Breast Height (DBH) | Entire 30 ∗ 30 m plot | Each resampling year | EBV: Terrestrial/Community composition/Community abundance; EBV: Terrestrial/Species populations/Species distributions | |
| Biodiversity | Forest | Dead tree inventories | Deadwood type, species name if possible, wood decay level, bark presence, wood volume | Entire 30 ∗ 30 m plot | Each resampling year | ||
| Biodiversity | Forest | Tree-related microhabitats inventory | Inventory of tree-related microhabitats on all living and standing dead trees with a DBH > 7.5 cm | Entire 30 ∗ 30 m plot | Each resampling year | Optional | |
| Biodiversity | Below-ground | Soil environmental DNA | Soil biodiversity characterization, using two universal markers (eukaryote and bacteria) and four clade-specific markers (Fungi, Insects, Oligochaeta, Collembola) | 3 “topsoil” subplot of 2 ∗ 2 m per plot | Each resampling year | EBV: Terrestrial/Community composition/Community abundance & Interaction diversity | |
| Biodiversity | Large mammals | Camera-traps | Large mammal presence during the summer | 2 cameras per plot | Each resampling year | Optional | |
| Biodiversity | Sound producing animals (e.g., birds, arthropods, amphibians, mammals) | Passive acoustic | Acoustic diversity metrics, bird and invertebrate detection through artificial intelligence | 1 acoustic recorder per plot | Each resampling year, 1 mn every 15 mn | Optional | |
| Biodiversity | Soil macroinvertebrates | Soil monoliths | Species name at highest resolution possible | 4 monoliths 25 ∗ 25 cm (∗20 cm deep when feasible) | Each resampling year | Optional | EBV: Terrestrial/Species populations/Species distributions & Species abundances |
| Biodiversity | Soil macroinvertebrates | Pitfall traps | Species name at highest resolution possible | 6 pitfalls per plot | Each resampling year | Optional | EBV: Terrestrial/Species populations/Species distributions & Species abundances |
| Biodiversity | Paleoecology | Description of anthracological fossil contents in soil pits | 1 “pit” subplot outside each plot | Once per plot | Optional | EBV: Terrestrial/Species populations/Species distributions & Species abundances |
Appendix B Consortium ORCHAMP Version 10/2024
B.1. Institutions involved in ORCHAMP
ORCHAMP is a consortium gathering a large range of actors: national and regional park managers, botanical conservatory experts, natural area conservatory managers, association, researchers from universities and research institutions. The project is led by the LECA (Laboratoire d’Écologie Alpine), located in Grenoble.
For additional information please visit our website: https://orchamp.osug.fr/home or contact us: orchamp@univ-grenoble-alpes.fr
- LECA - Laboratoire d’Écologie Alpine; Univ. Grenoble Alpes, Univ. Savoie Mont Blanc, CNRS, LECA, F-38000 Grenoble, France
- EDYTEM - Environnements, DYnamiques et TErritoires de la Montagne; Univ. Savoie Mont Blanc, Univ. Grenoble Alpes, CNRS, EDYTEM, Chambéry, France
- INRAE LESSEM - Laboratoire Ecosystèmes et Sociétés En Montagne; Univ. Grenoble Alpes, LESSEM, INRAE, Grenoble, France
- IMBE - Institut Méditerranéen de Biodiversité et d’Écologie marine et continentale; Aix Marseille Univ, Avignon Univ, CNRS, IRD, Marseille, France
- Jardin du Lautaret - Univ. Grenoble Alpes, CNRS, Jardin du Lautaret, F-38000 Grenoble, France
- CEFE - Centre d’Ecologie Fonctionnelle et Evolutive; Univ. Montpellier, CNRS, EPHE, IRD, Montpellier, France
- CEN - Centre d’Études de la Neige; Univ. Grenoble Alpes, Université de Toulouse, Météo-France, CNRS, Centre National de Recherches Météorologiques, Grenoble, France
- INRAE ECODIV - Laboratoire Etude et Compréhension de la bioDIVersité; Univ. Rouen - Normandie, INRAE, ECODIV, Rouen, France
- Laboratoire de Géologie de l’ENS; CNRS, ENS, Univ. PSL, Paris, France
- INRAE URFM - UR Ecologie des Forêts Méditerranéennes; INRAE, Avignon, France
https://ecologie-des-forets-mediterraneennes.paca.hub.inrae.fr/
- INRAE UEFM - UE Entomologie et Forêt Méditerranéenne; INRAE, Avignon, France
- DYNAFOR - Dynamiques et Écologie des Paysages Agroforestiers; INRAE, INP, Castanet Tolosan, France
- SETE - Station d’Ecologie Théorique et Expérimentale; Univ. Paul Sabatier, CNRS, Moulis, France
- CRBE - Centre de Recherche sur la Biodiversité et d’Environnement; Université de Toulouse, IRD, INP, CNRS, Université Toulouse 3 Paul Sabatier (UT3); Toulouse, France
- GEODE - Géographie de l’Environnement; Maison de la recherche Univ. Jean Jaurès, CNRS, Toulouse, France
- PatriNat - Centre d’expertise et de données sur le patrimoine naturel; OFB, MNHN, CNRS, IRD, Paris, France
- AR+I - Andorra Recerca + Innovació, Sant Julià de Lòria, Andorre
- LEM - Laboratoire d’Ecologie Microbienne; Univ. Lyon 1, CNRS, INRAe, VetAgro Sup, Lyon, France
- Eco&Sols - INRAE, IRD, CIRAD, Institut Agro, Montpellier, France
- ZA Alpes - Zone Atelier Alpes, Grenoble, France
- ZA PYGAR - Zone Atelier Pyrénées GARonne, Toulouse, France
- CBN Alpin - Conservatoire Botanique National Alpin, Domaine de Charance, 05000 Gap, France
- CBN Med - Conservatoire Botanique National Méditerranéen, Hyères, France
- CBN PMP - Conservatoire Botanique National Pyrénées et Midi-Pyrénées, Bagnère-de-Bigorre, France
- PN des Ecrins - Parc National des Ecrins & Réserve intégrale du Lauvitel, Gap, France
- PN du Mercantour - Parc National du Mercantour, Nice, France
- PN de la Vanoise - Parc National de la Vanoise, Chambéry, France
- PN des Pyrénées - Parc National des Pyrénées, Tarbes, France
- PNR du massif des Bauges - Parc naturel régional du massif des Bauges & Géoparc mondial UNESCO, Le Châtelard, France
- PNR de Chartreuse - Parc naturel régional de Chartreuse, 11 Place de la mairie, 38380 Saint-Pierre-de-Chartreuse, France
- PNR du Queyras - Parc naturel régional du Queyras & Réserve Naturelle Nationale de Ristolas - Mont Viso, 3580 route de l’Izoard, 05350 Arvieux, France
- PNR du Mont-Ventoux - Parc naturel régional du Mont-Ventoux, Carpentras, France
- RNR du Massif de Saint-Barthélemy - Réserve Naturelle Régionale du Massif de Saint-Barthélemy, Montségur, France
- RNR d’Aulon – CEN Occitanie, La Frênette et Commune d’Aulon
- Réserves Naturelles Catalanes - RNN de la vallée d’Eyne - Fédération des Réserves naturelles catalanes, 9 rue de Mahou 66500 Prades
- OFB - Office français de la biodiversité, gestionnaire de la Réserve nationale de chasse et de faune sauvage d’Orlu
https://www.ofb. gouv.fr/les-reserves/la-reserve-nationale-de-chasse-et-de-faune-sauvage-dorlu
- Observatoire de la montagne, Commune d’Orlu, Orlu, France
- Adyu l’Ome, Orlu, France
- ANA-CEN Ariège - Conservatoire d’Espace Naturel Ariège, Alzen, France
- Géoparc du Chablais - Géoparc mondial UNESCO du Chablais, Thonon-les-Bains, France
- Asters-CEN74 - Conservatoire d’espaces naturels de Haute-Savoie & Réserve Naturelle de Sixt - Fer à Cheval/Passy, Sixt-Fer-à-Cheval, France
- CREA Mont-Blanc - Centre de Recherches sur les Écosystèmes d’Altitude, Chamonix, France
- Natura 2000 Clarée - Névache, France
- Natura 2000 Dévoluy-Durbon-Charance-ChampsaurSMIGIBA - Syndicat Mixte de Gestion Intercommunautaire du Buëch et de ses Affluents Veynes, France
- Grenoble-Alpes Métropole - Grenoble, France
- ONF - Office National des Forêts - Grenoble
ag.isere@onf.fr
B.2. ORCHAMP Consortium (contact persons are in italics)
- LECA: Wilfried Thuiller, Amélie Saillard, Louise Boulangeat (2018–2021), Manon Bounous (2018), Irene Calderon-Sanou, Philippe Choler, Camille Desjonquères, Arnaud Foulquier, Ludovic Gielly, Priscilla Godfroy (2017–2019), Romain Goury, Maya Guéguen, Nicolas Le Guillarme, Clément Lionnet (2018–2023), Chloé Mahieu, Camille Martinez-Almoyna, Marc Ohlmann (2016-deceased in 2023), Gabin Piton (2016–2019), Julien Renaud, Matthias Rohr, Guillaume Terpereau (Student 2021), Tristan Ubaldi (Student 2019)
- EDYTEM: Jérome Poulenard, Nicolas Bonfanti, Norine Khedim (2018–2022), Emmanuel Malet, Lise Marchal (2019–2023), Erwan Messager, Yves Perrette
- INRAE LESSEM: Georges Kunstler, Vincent Breton, Laurent Berges, Nathan Daumergue, Adeline François, Sophie Labonne (retired in 2023), Laureline Leclerc (Student 2022), Eric Mermin, Jean-Matthieu Monnet (2018–2019), Yoan Paillet, Mathias Pires, Pascal Tardif (2016-retired in 2023)
- IMBE: Frédéric Guiter, Lenka Brousset, Cécile Albert, Armin Bischoff, Manuel Cartereau, Cécile Chemin, Emmanuel Corcket, Amandine Gasc, Raphaël Gros, Frédéric Guibal, Frédéric Médail (2018–2019), Eric Meineri, Jean-Philippe Mévy, Alexandre Millon (2018–2020), Pascal Mirleau, Daniel Pavon, Yoann Pinguet, Hervé Ramone, Caroline Rocher, Arne Saatkamp, Brigitte Talon
- Jardin du Lautaret: Jean-Gabriel Valay, Jérôme Forêt (since 2023), Rolland Douzet, Lucie Liger, Maxime Rome, Pascal Salze
- CEFE: Jean-François David, Cyrille Violle
- CEN: Samuel Morin, Matthieu Vernay, Matthieu Lafaysse
- ECODIV: Lauric Cécillon (2016–2022)
- Laboratoire de Géologie de l’ENS: Lauric Cécillon (2016–2022), Laure Soucémarianadin (till 2022)
- INRAE URFM: Bruno Fady, William Brunetto, Florence Courdier, Frédéric Jean, Nicolas Mariotte
- INRAE UEFM: Jean Thévenet, Marianne Corréard
- DYNAFOR: Laurent Larrieu, Antoine Brin, Laurent Raison, Célia Sirami, Catherine Bonnet, Jérôme Willm, Alexis Carteron
- SETE: Maxime Cauchoix
- CRBE: Jérôme Murienne, Gabrielle Martin, Renan Destrade, Uxue Suescun
- GEODE: Marie-Claude Bal, Mélanie Saulnier
- PatriNat: Olivier Delzons, Philippe Gourdain, Aurélie Lacoeuilhe
- AR+I: Benjamin Komac
- LBBE: Vincent Miele
- LEM: Juliana Almario, Lauren Gillespie
- Eco&Sols: Mickaël Hedde, Matthias Brand, Thomas Gelis, Nicolas Hénon, Cyril Versavel, Luna Vion-Guibert
- ZA Alpes: Mathilde Ratouis, Isabelle Arpin (till 2024), Renaud Jaunatre (since 2024), Marc Langenbach (since 2024), Erwan Messager (since 2024), Tamara Münkemüller (since 2024), Jérôme Poulenard (till 2024)
- ZA PYGAR: Arnaud Elger
- CBN Alpin: Bertrand Liénard, Sylvain Abdulhak, Léa Bizard, Gilbert Billard, Pauline Debay, Luc Garraud, Thomas Legland, Baptiste Merhan, Mathieu Michoulier, Gilles Pache, David Paulin, Thomas Sanz, Jérémie Van Es
- CBN Med: Virgile Noble, Pauline Bravet, Benoît Offerhaus, Henri Michaud, Maëlle Le Berre, Mathias Pires (till 2023), Julien Ugo, Marion Girardier
- CBN Pyrénées et Midi-Pyrénées: Jocelyne Cambecèdes, Ludovic Olicard, Michaël Douette, Anne Paris, Gilles Corriol
- PN des Ecrins & RI du Lauvitel: Richard Bonet, François Couilloud, Cédric Dentant, Damien Combrisson, Yoann Bunz, Jérôme Forêt (based in Jardin du Lautaret since 2023)
- PN du Mercantour: Clémentine Assmann (since 2024), Sébastien Honoré, Mathieu Krammer, Benoit Labigand, Marie-France Leccia (till 2024), Jérôme Mansons, Nathalie Siefert (till end 2023)
- PN de la Vanoise: Vincent Augé, Joël Blanchemain, Anne Bello (since 2024), Thierry Delahaye, Nicolas Gomez (since 2024), Franck Parchoux (2017–2021)
- PN des Pyrénées: Pierre Lapenu, Olivier Jupille, Jérémy Bauwin, Nils Paulet, Océane Pasquet, Sylvain Rollet,
- PNR du massif des Bauges: Jean-François Lopez, Richard Cousin
- PNR de Chartreuse: Bastien Moisan, Laure Belmont, Jessica Bruggeman (2019–2022)
- PNR du Queyras & RN Ristolas-Mont-Viso: Anne Goussot, Pierpaolo Brena, Alain Bloc (retired in 2023), Nicolas Tenoux
- PNR du Mont-Ventoux: Baptiste Montesinos, Anthony Roux
- RNR du Massif de Saint-Barthélemy: Laurent Servière
- RNR d’Aulon: Cyril Marmoex, Loyann Boy, Maëlle Benureau, Coline Carré, Lucyna Loriot (student 2024)
- Réserves Naturelles Catalanes - RNN de la vallée d’Eyne: Josep Parera Casas, Céline Quelennec
- OFB - gestionnaire de la RNCFS d’Orlu: Xavier Rozec
- Observatoire de la montagne, Commune d’Orlu: Christophe Lhez
- Adyu l’Ome: Pierre Guiton, Thérèse Sabadie
- ANA-CEN Ariège: Laurent Servière
- Géoparc du Chablais: Sophie Justice
- ASTERS & RN Sixt-Passy: Carole Birck, Olivier Billant, Jean-José Richard-Pomet
- CREA: Anne Delestrade, Bradley Carlson (2017–2023), Colin Van Reeth, Jeremy Froidevaux (since 2024)
- N2000 Clarée: Laure Vuinée
- Natura 2000 Dévoluy-Durbon-Charance-Champsaur & SMIGIBA: Eric Hustache
- Grenoble-Alpes Métropole: Alexandre Mignotte, Pierre-Eymard Biron (retired in 2021), Yann Kohler
- ONF: Carole Desplanque, Laurent Lathuillière
- Indépendants: Jean-Marie Dupont (Apexe), Françoise Laigneau, Christophe Perrier (till 2020), Olivier Senn, Alexandre Pailhé-Belair (Berger, estive d’Aulon), Alexis James (Berger, estive d’Aulon)
B.3. ORCHAMP funding
Each institution involved in the consortium is co-funding the project either through in-kind funding or participation to specific projects.
ANR - Agence Nationale de la Recherche: GlobNets (ANR-16-CE02-0009), Origin-Alps (ANR-16-CE93-004), TransAlps (ANR-16-CE02-0009) & EcoNet (ANR-18-CE02-0010-01)
ANR “Investissement d’Avenir”: Trajectories (ANR-15-IDEX-02), Montane (OSUG@2020: ANR-10-LAB-56), Idex UGA (ANR-15-IDEX-02); MIAI@Grenoble Alpes (ANR-19-P3IA-0003)
AFB - Agence Française pour la Biodiversité: Sentinelles des Alpes 2018–2019
OFB - Office français de la biodiversité: Sentinelles des Alpes 2020–2022, Sentinelles des Alpes 2023, Sentinelles des Alpes 2024
AURA - Région Auvergne-Rhône-Alpes: CBNA regional convention
SUD-PACA - Région Sud Provence-Alpes-Côte d’Azur: Support to CBNA and CBNMED
LTSER ZAA - Zone Atelier Alpes (CNRS, INRAE; membre de eLTER)
ISÈRE - Le Département: Appel à projets Biodiversité 2020
OSUG - Service d’observation: Appel à projets 2020
ECCOREV - Ecosystèmes Continentaux et Risques Environnementaux: Appel à projets 2019
Interreg Alcotra FEDER - PITEM Biodiv’ALP 2019-2023 PS3. - GEBIODIV
Interreg POCTEFA: FLORAPYR3D 2024–2026
BIOSEFAIR INRAE: SICCCUB 2021–2023
Région Occitanie: PAACTe Région Occitanie, 2022–2024
Interreg Alcotra FEDER: PITEM Biodiv’ALP
CNRS initiative EC2CO - Projet Microphos AAP 2021–2023
Other local fundings: Parc National des Ecrins (PNE), Réserve intégrale du Lauvitel, Parc National du Mercantour (PNM), Grenoble Alpes Métropole, Agence de l’eau Rhône-Méditerranée Corse (AERMC), Electricité de France (EDF), Mairie du Dévoluy, Institut de Radioastronomie Millimétrique (IRAM), Communauté de communes de la vallée de Chamonix Mont-Blanc, Fonds européen agricole pour le développement rural (FEADER).




 CC-BY 4.0
CC-BY 4.0