1. CNS regeneration: an unmet need
In adult mammals, central nervous system (CNS) neurons fail to regenerate their axon spontaneously, in contrast to the ones from the peripheral nervous system (PNS). This has major consequences for patients affected by neurological disorders, whether chronic (neurodegenerative diseases) or acute (traumatic brain or spinal cord injuries), as this leads to permanent and irreversible loss of motor, sensory and/or cognitive functions. To date, no treatment is available, making CNS repair one of the biggest challenges in Neuroscience research.
At the cellular level, when a CNS axon is lesioned, its distal part degenerates while the proximal forms a retraction bulb, a structure unable to bear axon growth. Among the reasons for this regeneration failure, the extracellular environment becomes growth-inhibitory upon injury, due to the release of growth-repulsive factors such as myelin-associated molecules or components of the glial border [1, 2, 3]. Yet, modulation of these inhibitory factors is not sufficient to promote robust regeneration [4], so researchers focused on the intrinsic growth properties of neurons themselves. Indeed, over the course of development, CNS neurons progressively lose the capacity to grow an axon [5]. Moreover, the injury itself further impairs the regrowth properties of adult CNS neurons [6], which correlates with the intrinsic downregulation of pro-growth molecular pathways. Over the past 15 years, axon regrowth of injured CNS neurons has been achieved through experimental activation or co-activation of these pathways, for example mTOR (mechanistic target of rapamycin) [7], JAK/STAT [8], Klf (Krüppel-like factors) [9] or c-myc [6].
Many of these breakthrough studies are based on the adult mouse visual system, as it is part of the CNS. It is composed of the eyes, the optic nerves and different brain nuclei that integrate and process visual information. The axon lesion is induced mechanically by crushing the optic nerve. This injury primarily affects retinal ganglion cells (RGCs), which are the only retinal neurons projecting their axon to form the optic nerve and connecting the eye to the brain (Figure 1).
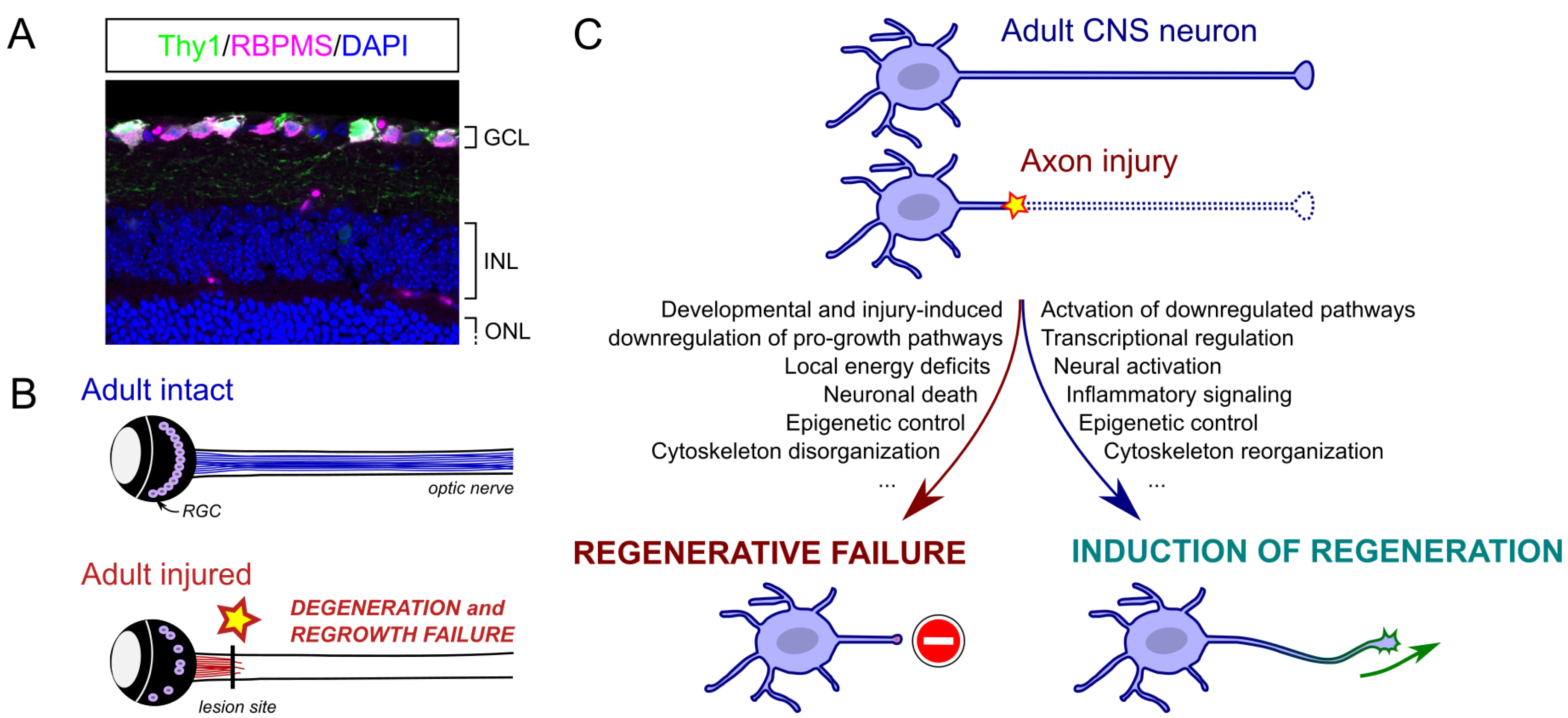
(A) Section of an adult retina showing specific labeling of RGCs with specific expression of GFP under Thy1 promoter and anti-RBPMS antibody. GCL: ganglion cell layer; INL: inner nuclear layer; ONL: outer nuclear layer. From [10]. (B) Schematic of optic nerve injury model. In normal conditions, RGCs project their axon via the optic nerve to the brain. After optic nerve injury, axons degenerate distally and fail to regrow. (C) Summary of molecular mechanisms regulating the neuronal injury response and the intrinsic regenerative properties of adult CNS neurons. Masquer
(A) Section of an adult retina showing specific labeling of RGCs with specific expression of GFP under Thy1 promoter and anti-RBPMS antibody. GCL: ganglion cell layer; INL: inner nuclear layer; ONL: outer nuclear layer. From [10]. (B) Schematic of optic ... Lire la suite
However, despite great advances in the field, many caveats remain. First, not all neurons are responsive to such molecular manipulations, partly owing to the heterogeneity in neuronal subpopulations. In the adult retina, more than 30 subpopulations of RGCs have been characterized, based on morphology, molecular markers, dendritic arborization, primary projections [11], and more recently transcriptional programs that condition RGC regeneration and survival after injury [12, 13]. Yet, modulation of such transcriptional programs is not sufficient to promote robust regeneration, as it remains unclear how they are integrated in the different subpopulations. Even the most robust models of regeneration enable no more than 20% RGCs to regrow, which might complicate to move to the next step: functional reconnection.
Second, regrowing RGCs fail to reach their proper target. Therefore, no functional circuit has been obtained yet [14, 6, 15]. This is due to the disorganized regrowth of regenerating axons, which do not resume their initial trajectory and cannot reinnervate their correct brain target [16, 17]. In this regard, our recent work has provided insight into the possibility and relevance of axon guidance modulation during regeneration, in order to reroute regrowing axons onto the correct path [18] and into the correct brain target [19].
Third, the molecular pathways identified to promote adult regrowth, such as mTOR, JAK/STAT and c-myc, have potential side effects. These pathways activate multiple cellular and molecular processes. This explains how axon regrowth can be obtained in the adult CNS, but this also accounts for the potential oncogenicity of such manipulations. Therefore, experimental strategies to promote axon regeneration are strong proof-of-concepts, but their application as a therapy has to be considered with caution. This is the case for the tumor suppressor Pten, whose inhibition in specific neurons in mice does not seem to cause detrimental effects in the long-term [20], but whose wide spectrum of targets may actually cause profound cell changes, and whose manipulation should be considered at multiple levels (gene expression, functional dosage, phosphatase activity, post-translational modifications, etc.) [21, 22].
Nonetheless current strategies remain a valuable experimental paradigm to elucidate the molecular mechanisms that control CNS regrowth capacity. Elucidating such mechanisms is fundamental to trigger and sustain axon regrowth, and ultimately address all subsequent steps of circuit repair that are just starting to be looked at, e.g., axon guidance in the mature brain as in our recent proof-of-concept studies [18, 23, 19]. So looking at these models will help understand how such pathways operate to promote axon regrowth and also neuron survival (as these two processes may be independent [24, 25, 26]), and refine what are the downstream targets that are necessary and sufficient for regeneration. Therefore, there is a critical need to find out other molecular mechanisms controlling axon regeneration in order to achieve full circuit formation and functional recovery.
2. Translational regulation at play to control gene expression
Until now, most studies have focused on transcriptional regulation to unlock adult CNS regenerative properties. In the adult visual system, pro-growth transcriptional programs were highlighted thanks to high-throughput screening, via RNA microarrays [27] and RNA-sequencing [26], and more recently via single-cell analysis of RGCs [28, 12, 13]. Another layer of gene regulation lies in epigenetic mechanisms at play to control expression of regeneration-associated genes (RAG). Indeed, regenerative transcriptional programs are governed by a modification of chromatin accessibility of RAG regulatory regions in response to injury, for example via regulation of histone acetylation status [29].
Transcriptomic analysis can stamp different types of cells according to detailed mRNA expression. This is the case in the retina, where high-resolution single-cell analyses have discriminated several dozens of neurons, during development and throughout adulthood [30, 31, 32, 33]. Yet, in many cases, transcriptome characterization does not account for functional or phenotypical properties of distinct subpopulations—and conversely indistinguishable subpopulations may actually harbor distinct transcriptomes [34]. This phenomenon is even more complex in the case of pathological or traumatic situations. Regarding axon injury and regeneration, modulation of transcriptional programs has been insufficient to recapitulate axon regrowth and neuronal circuit reconnection. So transcriptomic approaches are insufficient to predict major gene regulation processes in injured adult neurons.
In contrast, the process of mRNA translation into proteins—the functional readout of gene expression in cells—has been overlooked in the context of CNS injury and axon regeneration. While the activation of mTOR [7, 35] and c-myc [36, 6] pathways enhances protein synthesis at a global level—directly or indirectly—their exact contribution to translational control in this context remains unknown.
In fact, it is now well described that mRNA and protein levels correlate only partially in cells [37, 38, 39, 40] and in many cases, the transcriptomic characterization of a cell does not reflect the actual protein expression at a given stage. While translation was long described as an invariable process, evidence has recently emerged that pools of mRNAs are not evenly translated into proteins. Translation regulation mechanisms include differential activity of translation-associated factors [41], selective retention or degradation of mRNAs [41], and more recently variations in the composition of the translational complex itself [42].
3. Evidence for a specialized ribosome: focus on the translational complex
In eukaryotic cells, the translational complex is primarily composed of the ribosome, a macromolecular complex formed of about 80 ribosomal proteins and 4 ribosomal RNAs (rRNAs), which assemble in a tightly ordered process called ribogenesis. Polypeptide chain extension operates via screening of a given mRNA by the ribosome and translation of codon sequences by transfer RNAs into amino acids. This process also involves translation-associated factors that control the initiation, elongation and termination steps [43] (Figure 2).
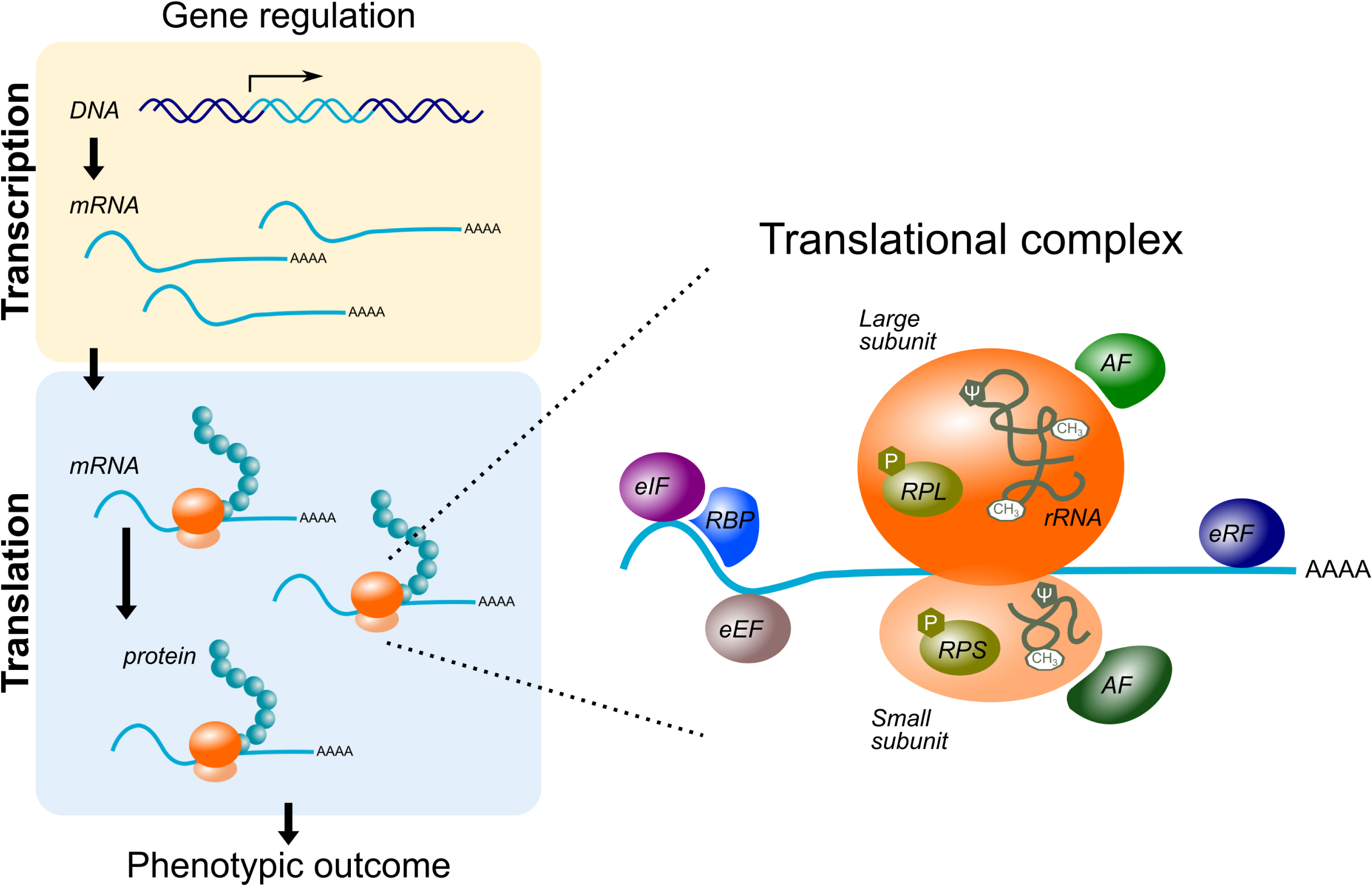
Schematic representation of gene expression control in cells: the genetic information flow goes from DNA to mRNA (transcription), then from mRNA to protein (translation). While long thought to be a passive process, translation is emerging as an active regulator in the control of gene expression. In particular, in pathological contexts, but also for cell specification, function and external stimulus-induced response (phenotypic outcome), the translational complex exhibits variations in its composition. These variations are associated with a specialization of its translational function. eIF, eEF and eRF: eukaryotic initiation, elongation and release factor; RBP: RNA-binding protein; RPL and RPS: ribosomal protein of the large and small subunits; rRNA: ribosomal RNA; CH3: methylation; 𝜓: pseudourydylation. Masquer
Schematic representation of gene expression control in cells: the genetic information flow goes from DNA to mRNA (transcription), then from mRNA to protein (translation). While long thought to be a passive process, translation is emerging as an active regulator in ... Lire la suite
While long thought to be invariable in its composition, the translational complex actually exhibits variations in its components and composition, and through them actively regulates the translation process in cells. One fundamental illustration lies in the hundreds of 2′-O methylation and pseudourydylation modification sites in rRNAs. Work from Diaz and colleagues shed light on how dysregulation of these modifications contributes to tumorigenesis by altering translation fidelity and mRNA-specific translation [44, 45, 46]. Another illustration lies in modifications of ribosomal proteins (RPs): either RP post-translational modifications or RP stoichiometry in ribosomes. These observations came from the study of so-called “ribosomopathies”, where somatic point mutations affect an RP or a ribogenesis factor. This leads to alterations in ribosome homeostasis and/or impaired translational regulation, and results in pathogenic outcomes, e.g., Diamond Blackfan anemia but also cancer development [47].
In physiological conditions, transcriptomic studies highlighted the existence of RP paralogs that confer tissue specificity and compensate their canonical counterparts. Examples include the ribosomal protein L3-like (Rpl3l), which is specific to the skeletal muscle and controls myotube formation [48]; and the ribosomal protein L10-like (Rpl10l), which is specific to the testis and controls male meiotic transition [49]. These paralogs are thought to replace their canonical counterparts in the ribosome, supporting further that the composition of the translational complex itself controls the translational landscape in tissue specification, development and function.
These studies pioneered the concept of “ribosome specialization”, a new and active field of research. Recently, RP heterogeneity in the composition of purified ribosomes was evidenced using quantitative proteomics and linked to a specialized function as a translation regulator [50]. Bulk proteomic analysis of various adult mouse tissues and organs also supports this inter-tissue heterogeneity at the ribosome-incorporated RP level [51, 52, 53]. Interestingly, in neurons, several pieces of evidence point towards an in situ remodeling of ribosomes by ribosome integration of newly locally synthesized RP in distal compartments of the neuron [54, 55]. This suggests an additional layer of control of the translational complex composition in subcellular compartments.
Finally, an additional level of heterogeneity comes from the translation-associated factors themselves. While the canonical translation factors are known to regulate the translation process at a global level [41], recent evidence is brought towards a role in selective (mRNA-specific) translation. For example, in Shh gradient-dependent patterning of the embryo, the initiation factor Eif3c controls Shh signaling through selective translation of Ptch1 mRNA, a negative regulator of Shh [56].
In addition, in different contexts, some non-canonical translation factors have been evidenced to (i) interact with the ribosome, and (ii) modify its translational activity, at the global level but also in a selective (specific mRNA-dependent) way. Examples include FMRP, shown to translationally repress target mRNAs linked to synaptic transmission [57]; and Rack1, shown to increase specific autophagy-related mRNA translation [58]. These observations make such associated factors integral components of the translational complex and contributors to its heterogeneity and specialization in cellular function. Recently, Simsek and colleagues provided an exhaustive “ribo-interactome” in mouse embryonic stem cells. They identified more than 300 ribosome-associated proteins susceptible to customize the ribosome, thereby influencing gene expression in specific physiological and pathological conditions [59].
4. Translational regulation in CNS axon regeneration
Our recent work shed light on translational regulation as an essential process of CNS axon regrowth: we deciphered this process from the molecular mechanisms to the subsequent in vivo implications regarding axon regeneration. Using the gold-standard optic nerve injury model, we demonstrated that axon regrowth of adult RGCs is controlled by the selective translation of a specific pool of mRNAs critical for axon regeneration—instead of global translation, i.e., even protein synthesis in the cell [60]. This mechanism is regulated by the differential association of specific factors to ribosomes in response to a pathological situation, in this case RGC axon injury. In turn, this modulates the regeneration-associated translational landscape in RGCs, independently of transcriptional regulation [60]. This study is based on the specific example of the protein Huntingtin (HTT), identified as a hub of the neuronal injury response [6, 61], which is quite unexpected since Huntingtin is better known for its neurodegenerative role in Huntington’s disease when the htt gene is affected by a CAG-expansion mutation. In contrast, HTT’s physiological roles remain elusive.
In the adult retina, HTT is downregulated in RGCs upon optic nerve injury. This downregulation correlates with the decrease in the regenerative potential of injured adult RGCs, supporting a pro-regenerative role for HTT. To test this hypothesis, we adopted the strategy to delete HTT expression specifically in RGCs and analyze its impact on regeneration. As the wild-type RGCs do not regenerate, we deleted HTT in a context where axon regeneration has been enhanced. To this end, we used the mTOR activation through deletion of the mTOR inhibitor Pten, within RGCs. Strikingly, HTT deletion in adult RGCs completely switches off axon regeneration induced by mTOR activation, meaning that HTT is essential for adult axon regrowth after injury (Figure 3A).
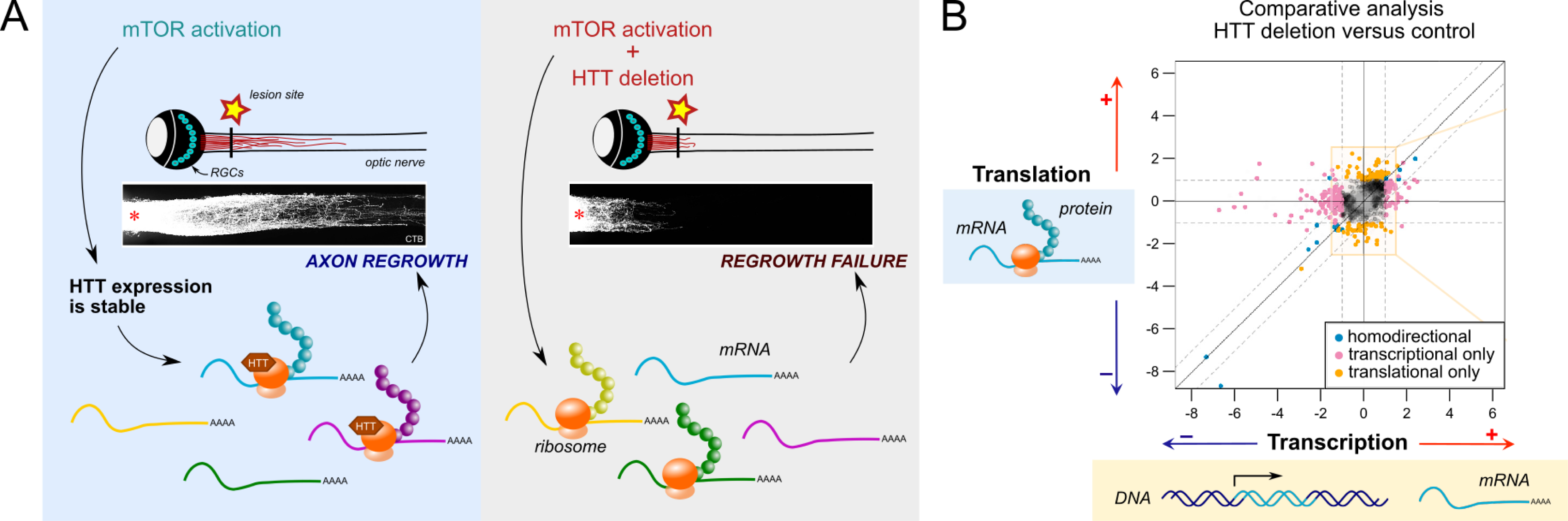
(A) Translational regulation is at play to control CNS axon regeneration. In regenerative conditions (mTOR activation), HTT expression remains stable upon injury. HTT interacts with ribosomes and regulates the translation of specific pro-regenerative mRNAs. When HTT is deleted, the translational complex is no longer customized with HTT. This modifies the pool of mRNAs recruited for translation and switches neurons to a regrowth failure state. (B) Comparative analysis of transcriptome and translatome in HTT deletion versus control conditions. Each dot represents an mRNA. The x-coordinate is its variation in transcript abundance (more or less transcribed when HTT is deleted). The y-coordinate is its variation in ribosome association (more or less translated when HTT is deleted). Orange dots represent the mRNAs that are selectively regulated at the translational level, with no variation of their transcriptional regulation. Among these orange mRNAs is Tox2, a pro-regenerative translational target of HTT. Data from [60]. Masquer
(A) Translational regulation is at play to control CNS axon regeneration. In regenerative conditions (mTOR activation), HTT expression remains stable upon injury. HTT interacts with ribosomes and regulates the translation of specific pro-regenerative mRNAs. When HTT is deleted, the translational ... Lire la suite
Starting from an unbiased analysis of HTT’s interactants, we showed that HTT interacts strongly with ribosomes, which suggests a role for HTT in the translation process. In vitro and in vivo new protein synthesis assays revealed that HTT is dispensable for global translation. In contrast, combined transcriptome and translatome analyses showed that, in the absence of HTT, a subset of mRNAs is differentially translated, while their total transcript level is unchanged. We focused on one such selective translational target of HTT, Tox2, to demonstrate that (i) HTT controls Tox2 translation in neurons, and (ii) the translation of Tox2 mRNA into protein is necessary for axon regeneration. This study elucidates, in the context of adult CNS injury, how ribosome association of specific factors, such as HTT, controls adult regrowth via translational regulation of a key subset of mRNAs [60] (Figure 3A–B).
Further evidence has been recently brought for the active role of the translational complex in the regulation of the translational landscape during axon regeneration. First, our work and others’ deciphered the essential role of kinases of the RSK family in the phosphorylation of the translation elongation factor eEF2 and of the ribosomal protein RPS6 [62, 63]. These studies demonstrate that RSK-mediated phosphorylation state is determinant for axon regeneration by controlling the translational landscape of specific mRNAs. Second, manipulation of one specific RP can also trigger RGC axon regrowth in the injured optic nerve, as shown for RPL7 and its isoform RPL7a whose transcripts are developmentally downregulated [64]. While this latter result further supports the role of the translational complex specialization in CNS regenerative response, it remains to be determined (i) whether experimentally overexpressed RPs integrate into functional ribosomes, and (ii) whether this confers an active translational regulatory role to the translational complex.
5. Perspectives
While long studied under the transcriptomic and epigenetic angles, the intrinsic regenerative capacity of the CNS depends on an additional layer of gene expression control: translational control through regulation of the translational complex. Such a mechanism has been exemplified by our recent work showing that HTT customizes ribosomes to regulate selective translation of regeneration-associated mRNAs in injured CNS neurons [60]. It is now more and more understood that transcriptomics alone cannot predict protein expression, not only through this active regulatory role of the translational complex, but also via accumulation and degradation mechanisms of individual mRNAs [41]. In this context, proteomic profiling should bring essential information to give a complete picture of how gene expression is regulated and how transcription, translation and proteostasis are integrated to control a phenotypic outcome, in this case CNS neuron injury response and axon regeneration. However, capturing the protein expression profiles in a single neuronal population, and a fortiori in single cells, remains a technical challenge capped by the current sensitivity of mass spectrometry-based proteomic analysis, as proteins cannot be amplified like nucleic acids. Yet, some studies could detect thousands of proteins in a single cell and pair transcriptomic and proteomic data [65], owing to major advances in shotgun proteomics such as SCoPE-MS [66] and PiSPA [67]. Such paired profiling provides highly valuable information on the regulation of gene expression at multiple levels. This can also apply to subcellular compartments, as performed recently in growth cones of developing projection neurons [68]. Today, translatomics and ribosome footprint profiling (deep-sequencing of ribosome-protected fragments) [69] offer a relatively accessible way to bridge transcription and protein synthesis levels, while effort will continue to improve the sensitivity of proteomic profiling.
Our findings are in line with a translational regulatory role for HTT. Indeed, in Huntington’s disease, HTT mutation was found to alter the translational process by impairing the activity of translation factors such as 4E-BP [70] or by impacting ribosome stalling [71]. Interestingly, we found a role in selective translation, while studies of mutant HTT show alterations in global translation. Thus, more investigation will be needed to decipher (i) which subdomain of HTT interacts with ribosomes (and in particular whether it is the N-terminal domain where the mutant poly-glutamine expansion is located); and (ii) whether HTT mutation affects its translational role directly or indirectly.
Whether such a mechanism of translational control applies to other neuron subtypes is a next question to address. In support of this hypothesis, we found that HTT is required for axon regeneration of dorsal root ganglion (DRG) neurons in the PNS upon sciatic nerve injury [60], and the work from others highlighted HTT as a key hub of regeneration of corticospinal tract motor neurons induced by neural progenitor cell grafting in the lesioned spinal cord [61]. Importantly, our work showed a correspondence in HTT-mediated control of the translational landscape between mouse embryonic fibroblasts and injured RGCs [60], suggesting a shared mechanism in a wide variety of cell types and developmental stages. In fact, HTT is ubiquitously expressed throughout multiple brain regions [72], and more generally in multiple tissues and at various developmental stages [73]. Additionally, studies of HTT in several model organisms advocate for a conserved role of its pathogenicity (induced by the mutated poly-glutamine expansion domain) and of its physiological functions, although these are still incompletely understood [74, 73, 75]. So it is possible that HTT’s role as a translation-associated factor is a common cell biology feature, but we cannot exclude that pools of translationally regulated mRNAs are actually specific to a given cell type, developmental stage or physiopathological condition, which will be worth investigating in the future.
What controls the selectivity of the translational complex to translate specific mRNAs? Some cis-regulatory elements identified in the sequence and/or structure of transcripts account for this selectivity. They include internal ribosome entry sites (IRES) in the 5′ untranslated region (UTR), upstream open reading frames (uORFs), 5′ terminal oligopyrimidine (5′ TOP) and miRNA-binding sequences [76, 77, 78, 79]. In CNS regeneration, it remains to be determined whether HTT translational targets contain such elements, and more generally whether there are consensus mechanisms of selective recruitment and translation of regeneration-associated mRNAs.
To investigate further the composition of the translational complex, an unbiased approach would be to determine with proteomics the ribo-interactome in neurons in response to axon injury, as recently done in mouse embryonic stem cells [59]. While working from neurons is quite challenging (low starting amount of proteins), advances in proteomic sensitivity should enable this characterization in the near future.
Finally, the translational regulation angle offers new perspectives in terms of therapeutic design for CNS repair. Our study led to the identification of HTT as a pro-regenerative factor, as well as its several translational targets whose experimental manipulation can promote CNS axon regrowth. Future work will focus on the modulation of translation-associated factors to regulate regeneration-associated mRNA translation.
To conclude, translational regulation is emerging as critical in the control of developmental programs, cell type specification, stress response [80, 42, 81], and now axon regrowth in the injured CNS [82]. Uncovering how the translational complex is customized in CNS neurons and how it contributes to translational regulation stands as a building step to fill the gap between gene expression and axon regrowth.
Declaration of interests
The authors do not work for, advise, own shares in, or receive funds from any organization that could benefit from this article, and have declared no affiliations other than their research organizations.
Funding
The work described in this review and their authors were supported by grants from the Agence Nationale pour la Recherche (ANR-18-CE16-0007 and ANR-21-CE16-0008-02), the Fondation pour la Recherche Médicale and UNADEV/AVISAN.
Acknowledgements
We thank all the authors involved in the work described in this review and the staff of Grenoble Institut Neurosciences facilities.





 CC-BY 4.0
CC-BY 4.0