1 Introduction
At the end of the 19th century, the concept of a mobile substance arose, which would be produced in leaves and move downward to induce root formation [1] or which would regulate the bending of grass coleoptiles toward the sun [2]. Later on, substances responsible for these phenomena were purified and characterized [3] and named “auxin” from the Greek “auxein”, literally “to grow/increase”.
The main natural auxin is indole-3-acetic acid (IAA) and is broadly found over evolution from bacteria to seed plants, including fungi, algae and even animals, from which the molecule was first chemically isolated [4].
Isolation of plant mutants related to auxin showed that modification of the regulation of auxin biosynthesis, transport or signalling generates severe alterations in many aspects of plant development. For example, the auxin over-producer mutant yucca leads to defects in vascular tissues formation [5]. Disruption in auxin transport, in the mutant pin1, leads to defects in floral development [6]. Finally, mutation in auxin signalling can trigger a global dwarfism as for the auxin resistant axr1.12 mutant [7], the absence of root formation as for the monopteros mutant [8], or even embryo lethality as for the abp1 null mutant [9]. This demonstrates that in plants, the phytohormone auxin plays a central role in plant growth and development.
Auxin is considered as a morphogen since it regulates development in a dose dependent manner [10]. It highlights the importance of auxin gradients and the necessity of a subtle regulation of auxin concentration at the scale of organ, tissues or even cells. To achieve such regulation, plants have developed various mechanisms aimed at controlling auxin homeostasis and the dynamics of auxin redistribution. In addition, various tissues exhibit distinct sensitivity to auxin, thus reflecting that the responsiveness (perception and signalling) is also tightly modulated.
The aim of this review is to provide a general overview on auxin and its involvement in development with a focus on primary root growth.
2 Auxin homeostasis
Auxin homeostasis results from multiple dynamic and continuous adjustments of regulatory mechanisms, aimed at maintaining a fairly stable internal equilibrium, allowing effective plant growth and development, and adaptation to a broad range of environmental stimuli. The mechanisms controlling homeostasis involve auxin biosynthesis, conjugation, catabolism, cellular compartmentalization and transport. Existence of multiple parallel auxin biosynthetic pathways provides a well-buffered and versatile system ensuring production and renewal of auxin whatever the conditions. Interestingly, active auxin transport is responsible for the establishment of auxin gradients within plant tissues, thus disturbing locally the homeostasis to trigger differential cellular responses, which are essential to sustain plant growth and development and to respond rapidly to environmental changes.
2.1 Auxin biosynthesis
IAA is mostly synthesized in young leaves [11] and then transported throughout the plant even though the importance of local IAA biosynthesis in specific tissues has been shown to be essential for plant development. Within the last decade, many advances have been made in the capacity to quantify free IAA and its metabolites from small amounts of plant tissues (less than a milligram of fresh weight), allowing refinement of the previous view of auxin distribution within plant tissues [12,13]. In addition, detailed analysis of auxin content combined with the use of transport inhibitors or auxin biosynthesis mutants has provided evidence supporting that auxin biosynthesis is not restricted to young leaves and can occur, at least under certain circumstances, in most tissues including roots [12,14]. More recently, root cell type-specific analyses of IAA content and metabolites, using fluorescence-activated cell sorting of GFP-specific cell markers combined with highly sensitive mass spectrometry methods, revealed an elevated capacity for IAA biosynthesis of almost all cell types at the root apex [15]. Plant IAA biosynthetic pathways are still incompletely elucidated, but the use of stable isotopes has revealed that IAA can derive from two major sets of pathways. One is known as the tryptophan (Trp)-dependent pathways and can use up to four distinct routes; the other derives from a precursor of Trp and is referred as the Trp-independent pathway (Fig. 1). Major features of IAA biosynthesis are summarized here, but this topic has been extensively reviewed by [16,17].
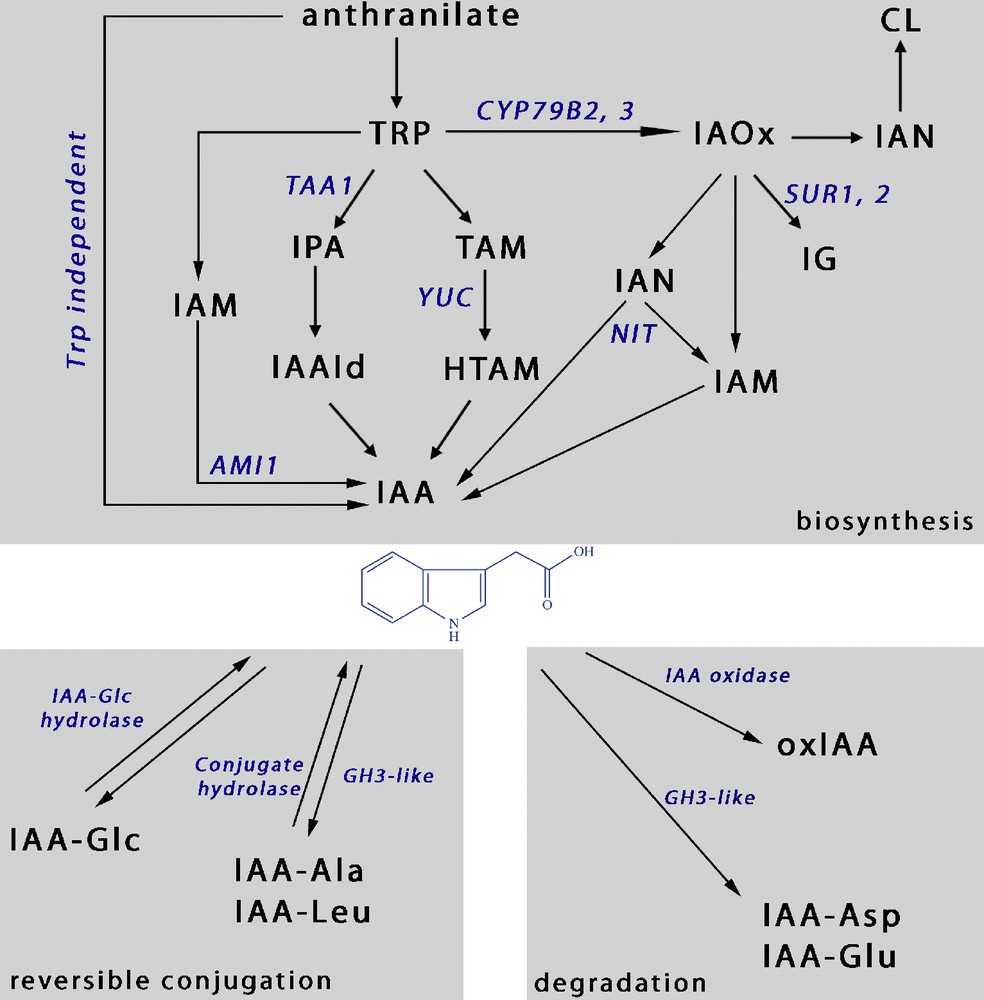
Auxin homeostasis, various routes for IAA biosynthesis and conjugation. The upper grey panel represents the known IAA biosynthesis pathways, including Trp-independent and Trp-dependent pathways. The lower panels illustrate the contributions of reversible and irreversible conjugation or oxidation in the control of auxin homeostasis.
2.2 Trp-dependent IAA synthesis pathways
Four different routes have been characterized from Trp to IAA, involving various intermediates to IAA (Fig. 1-biosynthesis). These pathways are not parallel and the routes often cross. One has indolacetamide (IAM) as an intermediate which is transformed to IAA by an amidohydrolase (AMI1). In another one, Trp is transformed into indole-3-pyruvic acid (IPA) by an aminotransferase (TAA1). Then, an IPA decarboxylase transforms IPA into indole-3-acetaldehyde (IAAId), which is then converted into IAA by an IAAId oxidase (AAOI, not on diagram).
One more of these pathways includes the transformation of Trp into tryptamine (TAM) by a Trp decarboxylase then the YUCCA (YUC) proteins function in the conversion of TAM to N-hydroxyl-TAM (HTAM) leading to IAA after further modifications [17,18]. Over expression of the YUCCA1 protein in the yucca mutant is sufficient to alter root development leading to a shorter and thicker primary root, demonstrating the importance of tight regulation of IAA production in the roots [19].
The last pathway is the most ambiguous one. Trp is converted into indole-3-acetaldoxime (IAOx) by two cytochrome P450 (CYP79B2/CYP79B3) proteins, IAOx is then converted into indole-3-acetonitrile (IAN) which is finally converted into IAA by nitrilases (NIT) expressed in roots [20]. The cyp79B2,cyp79B3 double mutant exhibits defects in root growth [21] and a decrease in local auxin production [12] demonstrating the existence and the importance of local IAA production in roots through this pathway. It is known that IAOx can also be transformed into 3-indolylmethyl-glucosinolate (IG), a metabolite that deters herbivores from eating the plants. IAA overproduction was reported in Arabidopsis superroot 1 and 2 (sur1, 2) mutants [22,23], which do not produce enzymes responsible for IG synthesis. Both mutants exhibit enhanced root formation as a consequence of the increased of IAA content, likely to result from a conversion of IAOx via NITs. A direct pathway allowing IAA biosynthesis from IG has recently been questioned at least in Arabidopsis [18].
2.3 The Trp-independent IAA synthesis pathway
A Trp-independent IAA synthesis pathway in plants is regularly questioned, as the detailed pathway has still to be elucidated. There are, however, obvious clues pointing towards the existence of this Trp-independent IAA biosynthesis (Fig. 1-biosynthesis), starting with the characterization of Trp deficient mutants. The arabidopsis trp3-1 and trp2-1 mutants which are defective in Trp synthase α and β, respectively [24,25], can still accumulate IAA conjugates despite low Trp content [26,27]. In these mutants, IAA might be produced from Trp precursors such as indole-3-glycerol phosphate or indole [16,17].
Much progress has been made in our understanding of auxin biosynthesis but much of our knowledge it is still fragmentary. The relative importance of the distinct biosynthesis routes in various tissues, plant species or growth conditions is poorly documented. Regulation of these biosynthetic pathways and cross talk with other plants signals have still to be further investigated.
2.4 Reversible IAA-conjugation or conversion
De novo biosynthesis is not the only source of free IAA. Higher plants can store IAA in the form of IAA conjugates and indole-3-butyric acid (IBA), which can provide free IAA upon hydrolysis or β-oxidation (Fig. 1). IAA can be linked to sugars, amino acids such as alanine (Ala) or leucine (Leu), or peptides [28] (Fig. 1). Proposed functions for these conjugates include storage, transport, compartmentalization, detoxification of excess IAA, and protection against peroxidative degradation [29].
In response to a specific signal or at specific times, cells may have to quickly reduce their inner auxin concentration, which can be rapidly achieved through conjugation. On the other hand, these reversible reactions allow transformation back to free IAA when the cell needs to release more auxin.
2.5 Irreversible conjugation and oxidation
Other conjugates such as IAA-Glutamate (Glu) and IAA-Aspartate (Asp) are also present in Arabidopsis [30,31]. Active auxin cannot be released by the hydrolysis of these compounds which are thus part of a catabolic process, IAA-Glu and IAA-Asp leading to degradation [32].
Remarkably, group II of GH3-like proteins, which have been shown in vitro to conjugate amino acids to IAA, seem to favor conjugation to Asp or Glu rather than forming hydrolysable IAA-Ala or IAA-Leu conjugates [33] (Fig. 1). Group II GH3 genes are primary auxin response genes. Elevated IAA levels induce their expression thus increasing a catabolic conjugation pathway as a way to maintain auxin homeostasis. Another possibility for the cell to decrease auxin concentration is to oxidate IAA into OxIAA (Fig. 1). This oxidation is known to be irreversible and to play a major role in the IAA catabolic pathway, thus contributing to auxin homeostasis [31,32].
3 Auxin transport
The hormone auxin, as a mobile signal, needs to be transported to accomplish its role of messenger between cells, tissues or organs. Auxin transport is for sure the most investigated topic in the “auxin field” nowadays and we have made considerable breakthroughs during the recent years.
As indicated above, auxin is mainly synthesized in young leaves and transported throughout the whole plant. There are two types of auxin transport, long distance transport and cell-to-cell auxin transport. Most IAA is transported via the phloem which confers a natural flow from the source tissues to the sinks. This long distance transport is efficient but cannot be finely regulated in comparison to the highly regulated polar auxin transport (PAT) between cells.
To understand cell-to-cell auxin transport we need to step back and look at auxin as a chemical molecule. Owing to its small size and its lipophilic characteristic, auxin can easily diffuse through the cell wall and cell membrane. Auxin is a weak acid (pKa = 4.75) and the proportion between the hydrophilic anionic and the hydrophobic protonated forms depends on the local pH. In the apoplast, the pH oscillates between 5.5 and 5.7 which is sufficient to have about 16% of IAA in the protonated form (IAAH), whereas in the cytosol, the pH is fairly stable around 7.4, favoring accumulation of the non-diffusible IAA anion (IAA-). This difference of pH favors the directional diffusion of the protonated IAA into the cell.
Even though auxin can passively enter the cell, it is clear that active transport uptake through specific carriers is important. Auxin influx carriers have been identified by the characterization of an Arabidopsis mutant, aux1, resistant to the synthetic auxin 2,4-dichlorophenoxyacetic acid (2,4-D) [34,35]. Diffusion of 2,4-D into the cell is poor and the uptake of this compound requires the activity of influx carriers. Interestingly, the aux1 mutant is not resistant to another synthetic auxin, naphthalene acetic acid (NAA), which is highly diffusible and does not make use of specific carrier to enter the cells [35]. AUX1 and three closely related genes named LAX (Like AUX1) belong to a subfamily of amino acid permeases [36]. The AUX1/LAX proteins act as symporters enabling entry of IAA− with the help of the proton gradient [37]. These influx carriers have specific roles in various processes such as root gravitropism [36] and lateral root formation [38] and are thought to be required to prevent dissipation of the differential accumulation of auxin, thus maintaining the local auxin flow [39].
Once auxin enters the cytosol, the IAA anion is unable to get out of the cell through diffusion across the plasma membrane. Its export was thus proposed to be assisted by a transmembrane efflux carrier [40] and the asymmetric cellular localization of these efflux transporters has been proposed to determine the direction of auxin flow [41].
Genetic analysis of Arabidopsis mutants with auxin-related/developmental phenotypes, resulted in the identification of two main gene families of auxin efflux carriers. The first discovered were members of the PIN family named after the pin formed 1 mutant, which exhibits strong defects in flowers or lateral organs along its inflorescence [6]. The same phenotype can be obtained by treating plants with an auxin transport inhibitor, the N-(1-naphthyl)-phthalamic acid (NPA) [6]. These proteins are predicted to contain 6 to 10 transmembrane domains, similar to bacterial transporters [42,43]. PIN proteins are expressed in tissues and cells transporting auxin and are asymmetrically localized within the cell at the plasma membrane. Polar cellular localization of PINs seems to be determined by auxin itself [44]. The model developed by Friml et al. presents PIN protein polar localization at one side of cells as the result of a continuous cycle of endocytosis and exocytosis [45]. Auxin was demonstrated to be a signal inhibiting PIN recycling by decreasing their endocytosis. The phosphorylation state of the PIN proteins seems to determine the directional exocytosis of the PINs. The PINOID kinase seems to favor accumulation on the apical side of the cell whereas protein phosphatase 2A favors accumulation at the basal side [46,47].
The PIN proteins have different patterns of expression and together form routes conducting auxin flow and local redistribution (Fig. 2). At the root meristem, PIN1, PIN3, PIN4 and PIN7 are basally (meaning the cell surface oriented towards the root apex) localized in the stele cells, leading auxin flow toward the quiescent center (QC) close to the root tip [48,49]. A change in auxin flow from the columella to the lateral root cap and the epidermis is achieved by the lateral localisation of PIN3 and PIN7 [50,51]. On the other hand, PIN2, which is located on the apical side of epidermal cells, redirects auxin upward to the end of the meristematic zone in which PIN1, PIN3 and PIN7 recycle auxin to the stele to close the loop of auxin flow.
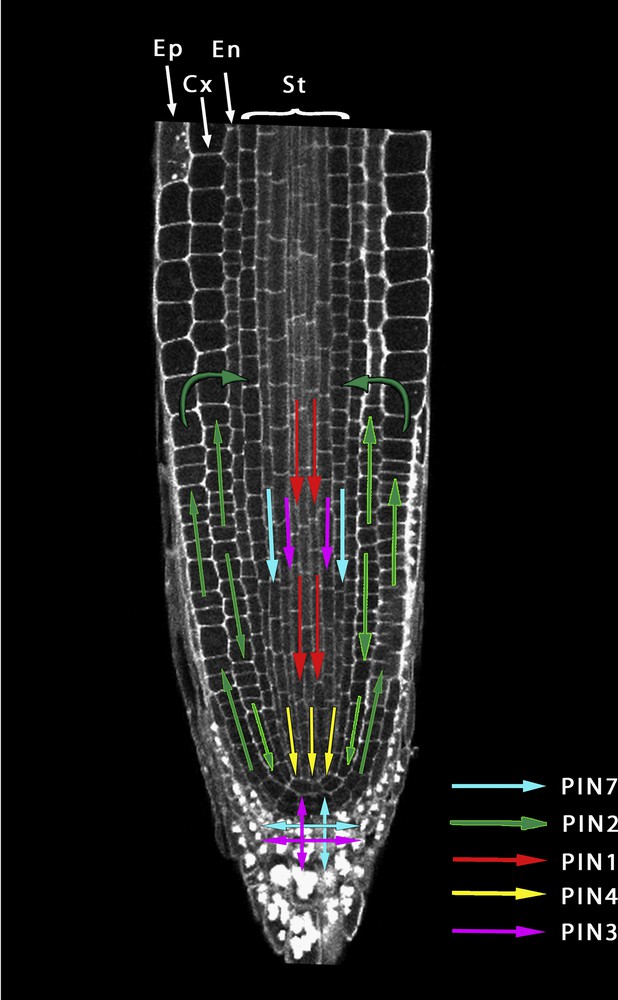
Visualization of auxin fluxes generated by the various PIN auxin efflux carriers at the root apex. Polar localization of PIN proteins in the respective cell files sustains high auxin levels around the quiescent center and within the root meristem and a reduction of auxin at the transition zone through a dynamic recycling of auxin. Ep: Epidermis, Cx: Cortex, En: Endodermis, St: Stele tissues.
Several other proteins demonstrated to be involved in auxin efflux belong to the multigene family of ATP-Binding Cassette (ABC) transporters also known as multidrug resistant proteins or P-glycoproteins (MDR-PGPs) [52,53]. These proteins exhibit auxin transport activity when expressed in a heterologous host [54] and were identified as targets of the transport inhibitor NPA [55]. Furthermore, the mutants mdr1 and mdr1,pgp1 exhibit severely reduced PAT [52]. The importance of these MDRs for auxin related development has been reported in other higher plants [56]. In the root, MDR-PGP transporters are proposed to support auxin flow mediated by PINs. For example, ABCB1 is expressed in all root cells except for the columella, whereas ABCB19 expression is restricted to the endodermis and the pericycle which might help to separate auxin fluxes toward the root tip through the stele and from the root tip to the epidermis [57,58].
Both auxin influx and efflux transport are indispensable to proper root development from embryogenesis to meristem maintenance and formation of lateral roots. In the primary root, PAT allows recycling of auxin and constant renewal of the gradients. Root cell-type specific analyses of IAA content revealed the fine patterning of auxin distribution with high auxin concentration in the QC, less elevated concentration in the endodermis and cortex and weak concentration in the epidermis [15]. In epidermal cells, the auxin level decreases at the beginning of the elongation zone, just above the point of its recycling back to the stele [59] [60]. Changes in local auxin concentration are believed to determine cell fate during root growth. Similarly, PIN3 and PIN2-mediated asymmetric redistribution of auxin following gravistimulation inhibits cell elongation of epidermal cells on one side of the root and triggers the bending of the root [39,61].
In addition to its critical role in root development, a tight regulation of PAT is also essential for embryogenesis with the establishment of the embryo polarity, for phyllotaxy or vascular formation in leaves [56].
4 Auxin signalling
During the last decade, the field of auxin signalling made a big step forward with elucidation of at least part of the mechanism sustaining rapid modulation of gene expression in response to auxin.
A very short and efficient pathway involving an auxin receptor, located in the nucleus, has been identified. When auxin enters into the nucleus, it allows recognition and degradation of repressors of auxin-regulated gene expression. Once the repressors are degraded, transcription of early auxin response genes can occur directly and leads to auxin responses. Another protein, the auxin binding protein 1 (ABP1) has been shown to participate in auxin responses at the plasma membrane, to mediate auxin control of cell division and cell elongation, and lately, to modulate regulation of early auxin response genes supporting its presumed function as an extracellular receptor of auxin.
Surveys of cells after auxin treatment revealed that part of the auxin response is mediated by modification of gene expression and that it does not require de novo protein synthesis [62]. A combination of biochemical, molecular, and genetic approaches allowed the identification of three main families of early auxin response genes expressed within 5 to 60 minutes after auxin treatment [63]. These families are the Small Auxin Up RNA (SAURs) genes, the GH3s and the Auxin/IAA inducible genes (AUX/IAAs) [64]. SAURs genes code for highly conserved short-lived small transcripts [65] and although their function is not clearly established, they have been proposed to act as calmodulin-binding proteins [66]. GH3 genes code for conjugating enzymes, a class of which acts as feedback regulators by reducing free auxin levels [33]. Amongst the 29 genes of the AUX/IAA family in the Arabidopsis genome, most members are upregulated by auxin [63,67,68]. They encode short-lived transcriptional repressors of the auxin response genes, contributing to keeping their expression low in the absence of auxin and to exert a negative feedback on the expression of auxin response genes after auxin stimulus. AUX/IAA repressors are critical for transient and brief auxin mediated responses as discussed in [69].
Analysis of the promoter region of the IAA4/5 gene from the pea and a GH3 gene from the soybean [70,71] revealed a common sequence named Auxin Response Elements (AREs) with a consensus element TGTCTC. This cis element was shown to confer auxin inducibility to minimal promoters [72,73] and led to the construction of a number of synthetic auxin-responsive promoters such as DR5, which contains multimers of the TGTCTC sequence fused to a constitutive promoter element [74].
Yeast one hybrid analysis using the ARE as a bait allowed the identification of Auxin Response Factor1 (ARF1) [75]. ARF1 belongs to a family of 23 genes in the Arabidopsis genome [76]. Most ARFs possess a DNA binding domain at the N-terminal and are transcription factors involved in the regulation of early auxin response genes. In general, they act as activators if they contain a glutamine/serine/leucine-rich (QSL-rich) middle region or as repressors if they contain a serine or serine/proline/glycine-rich middle domain [77–79]. A number of ARFs exhibit two C-terminal protein-protein interaction domains referred as domains III and IV.
These domains III and IV are shared by the Aux/IAAs and may trigger homo- or hetero-dimerization with ARFs or the AUX/IAA repressors. In the absence of auxin or at low auxin levels, ARF and AUX/IAA heterodimers might be formed [80] (Fig. 3A). The N-terminal protein–protein interaction domain of the AUX/IAAs (domain I) can also interact with other transcriptional repressors to exert repression (Fig. 3A). TOPLESS (TPL) is an example of a co-repressor which can physically interact with the domain I of AUX/IAA12 (BODENLOS, BDL) [81]. TPL is required for BDL repressive activity on auxin response and the dominant negative tpl1 mutation suppresses the patterning defect of the bdl1 gain of function mutant. In two-hybrid experiments, TPL also interacts with other Aux/IAAs. TPL and the other members of its family act presumably as coregulators of auxin responses. Another example of combinatorial transcriptional control is the involvement of MYB77 [82]. This protein has been shown to interact with domain III and IV of ARFs in vitro. Experiments with protoplasts confirmed that the coexpression of MYB77 and an ARF C-terminus enhances reporter gene expression. PKL (PICKLE) is a corepressor involved in constitutive repression of auxin response by AUX/IAA14 (SOLITARY ROOT, SLR) [83,84]. PKL is a homologue of the animal chromatin-remodeling factor CHD3/Mi-2 and might be a subunit of a complex containing histone deacetylases. Experimental evidence suggests that PKL/SSL2-mediated chromatin remodeling negatively regulates auxin-mediated lateral root formation in Arabidopsis.
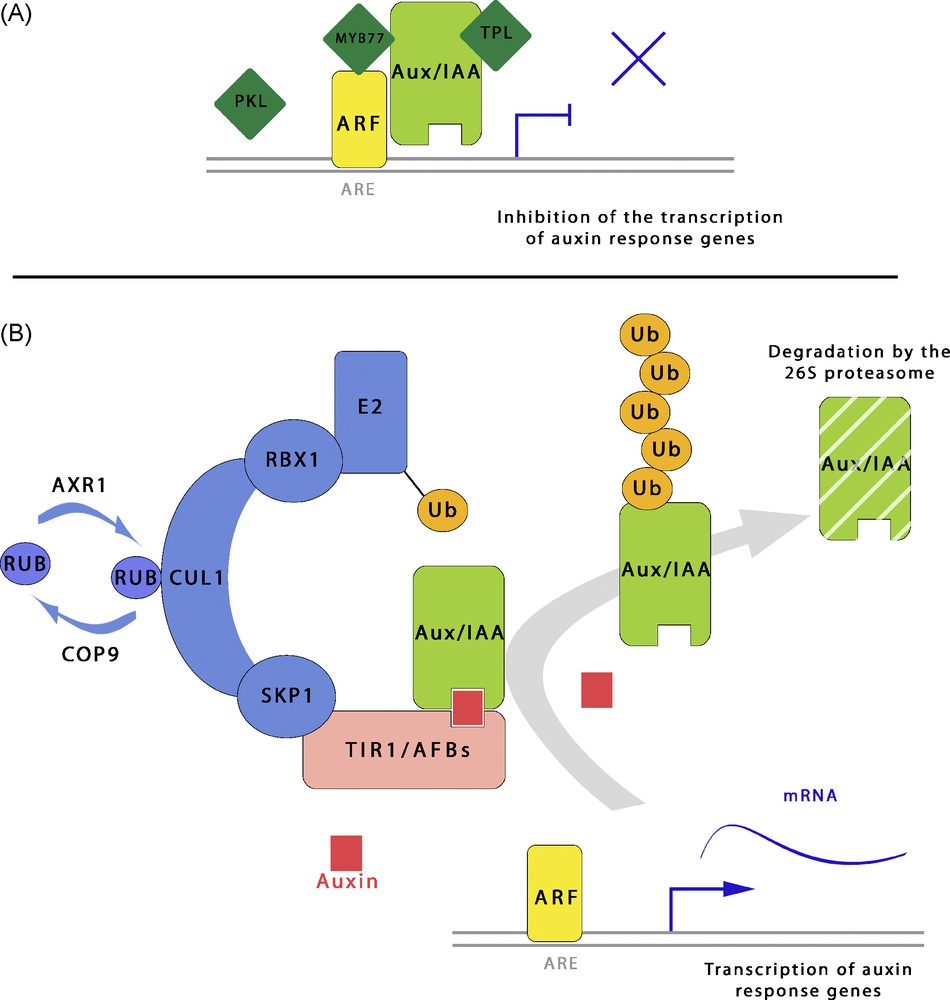
Simplified model of the auxin signalling pathway controlling the transcriptional regulation of early auxin response genes. (A) In the absence or at low level of auxin, a combination of transcriptional repressors inhibit the activity of AUXIN RESPONSE FACTORS (ARFs) which interact with auxin responsive elements (ARE) found in promoter domains. The AUX/IAAs repressors associate with ARFs which bind directly to the ARE. Other proteins can be associated with this transcription regulation, such as PICKLE (PKL) which participates in chromatin remodeling, the corepressor TOPLESS (TPL) or the positive regulator MYB77. (B) Elevated auxin level in the nucleus promotes degradation of the AUX/IAA repressors by the 26S proteasome. Auxin acts as a molecular glue between AUX/IAAs and TIR1/AFBs, subunits of the SCF complex which adds multiple ubiquitins to the AUX/IAA substrates, thus targeting them for degradation by the proteasome. This results in derepression of ARF activators, and transcription of auxin response genes. Masquer
Simplified model of the auxin signalling pathway controlling the transcriptional regulation of early auxin response genes. (A) In the absence or at low level of auxin, a combination of transcriptional repressors inhibit the activity of AUXIN RESPONSE FACTORS (ARFs) which ... Lire la suite
Indeed, plants have developed various ways to repress auxin responses, increasing the complexity of its regulation. In the general scheme of auxin response repression, it is essential to take into account the spatial and temporal regulation of the expression of both AUX/IAAs and ARFs, as well as the relative specificity of interactions between these transcription regulators. It is also likely that the distribution of AREs within promoters and their sequence environment may modulate the amplitude of the auxin response at those loci. Variation in ARE sequences and abundance may serve as the first level of complexity in the transcriptional regulation of auxin-responsive genes, as discussed in [85].
The understanding of the regulation of gene expression in response to auxin is based on the analysis of the regulation of representative members of three gene families, identified as early responsive genes. Recent large scale analysis of gene expression in response to auxin has revealed that short term auxin treatment results in changes in expression of hundreds of genes, indicating that many other regulators might be involved to the control of auxin responses [86,87].
Derepression of auxin responses occurs after an increase in the intracellular auxin level. Elevation of auxin in the nucleus promotes the targeted degradation of the AUX/IAA repressors by the 26S proteasome (Fig. 3B). Auxin acts as molecular glue increasing the interaction of the domain II of AUX/IAAs with Transport Inhibitor Response1/Auxin related F-Box (TIR1/AFBs), F-box proteins of the E3 ubiquitin ligase complex SKP1-CULLIN1-Fbox-TIR1/AFBs (SCFTIR1/AFBs). For these reasons, TIR1 is considered as an auxin receptor [88,89]. Recruitment of AUX/IAAs by the TIR1/AFBs F-box provokes the polyubiquitination of the AUX/IAAs substrate and its degradation by the 26S proteasome (Fig. 3B) [90–92]. TIR1 was originally identified from the transport inhibitor resistant 1 (tir1) Arabidopsis mutant [93], after which five closely related genes named AFB1 to 5, belonging the LRR-rich class of F-box-encoding genes, were proposed to act like TIR1 in auxin signaling [4,94]. The TIR1 F-box recognizes a consensus sequence GWPPV/I from domain II of AUX/IAAs which was shown to be sufficient to confer short half-life to the proteins, thus defining this motif as a degron [90]. Proper regulation of the half-life of AUX/IAAs is essential for auxin dependent plant development. Indeed, plants with a single mutation in an AUX/IAA degron accumulate the corresponding protein which can no longer be degraded [4]. Such gain of function mutants exhibit various developmental defects, such as axr2_1 (iaa7) which suffers from a defect in the root gravitropic response [95,96] or solitary root slr (iaa14) which cannot develop lateral roots [97]. Likewise, the bodenlos (iaa12 gain of function) mutant [8] or the monopteros (ARF5 KO mutant) [98], which have been demonstrated to work together to mediate auxin signalling in the embryo, exhibit a rootless phenotype pointing out the importance of proper auxin signaling during the formation of the root in the embryo. A rootless phenotype is also observed in one third of the population of tir1, afb1, afb2, afb3 quadruple mutants [94].
Little is known about the relative affinity of interaction between various AUX/IAAs and the different TIR1/AFBs F-Boxes. The binding of auxin was shown for TIR1 in the presence of AUX/IAA peptides, but it is not clear whether the F-box alone can bind auxin or if the complex formed with AUX/IAA is required for sequestration of auxin at the interface between the two proteins. The capacity of auxin binding may also vary from one interacting pair (an AUX/IAA and TIR1 or an AFB) to another. Albeit potential differences of expression in the different TIR1/AFB members, it would be interesting to determine whether the various TIR1/AFB proteins are fully functionally redundant.
Another way to regulate the expression of early auxin response genes is to modulate the quantity of F-box proteins as a result of the regulation of the amount of TIR1/AFBs mRNA by miRNA393 as demonstrated for TIR1, AFB2 and AFB3 in case of pathogen defense mechanisms [99].
Multiple subunits are involved in the activity of the E3 ubiquitin ligase SCF complex [131] including CULLIN1 (CUL1), a molecular scaffold protein bringing together the two functional parts of the SCF complex [100,101]. The catalytic module is formed by a RING finger domain protein (RBX1) and the E2 ubiquitin conjugating enzyme. The F-box protein providing the specificity of recognition of the substrate is included in the second domain of the SCF. The dynamics of assembly and disassembly of the complex requires modulation of a post-translational modification of the CULLIN1 through ligation of a single Related to Ubiquitin (RUB) protein. One of the first auxin resistant Arabidopsis mutants isolated in an auxin root growth screen was axr1 (auxin resistant 1) [102–104]. AXR1 codes for a subunit of the RUB-E1 activating enzyme which mediates the rubylation of CUL1. Conversely, derubylation is mediated by the multiprotein complex COP9 signalosome [105] (Fig. 3B).
The SCFTIR1/AFBs auxin signalling pathway is short and controls the auxin-induced changes of gene expression by targeting the degradation of transcriptional repressors. With the identification of various corepressors, and with the involvement of additional transcriptional factors participating in a combinatorial regulation of early auxin response genes, there are increasing question marks addressing whether the SCFTIR1/AFBs pathway is sufficient to tightly regulate auxin regulated gene expression. Recently, two additional proteins were shown to be involved in the regulation of auxin response gene expression. One is the long standing AUXIN BINDING PROTEIN1 (ABP1) receptor which is involved in very early auxin mediated responses at the plasma membrane and was recently reported to be necessary for the tight regulation of early auxin response genes in Arabidopsis [116,118]. The other is a dual-specificity phosphatase-like protein, named Indole 3-Butyric acid Response 5 (IBR5), which was reported to promote auxin responses through a pathway distinct from TIR1-mediated repressor degradation [127]. The SCFTIR1/AFBs pathway is a core component of the mechanism controlling auxin action on gene expression but it is also clear that it is not the sole auxin signalling pathway. One of the exciting challenges of the coming decade will be to identify other pathways and to integrate them all in a comprehensive model.
ABP1 was discovered owing to its capacity to bind auxin [106,107]. This protein is located at the plasma membrane (PM) and at the endoplasmic reticulum (ER) [108,109]. There is only one ABP1 gene in Arabidopsis and it is conserved in the whole plant kingdom even in green algae [110]. Its function as an auxin receptor at the PM was demonstrated by many experiments, notably using electrophysiology. Auxin responses at the PM, such as activation of the H+/ATPase pumps or modification of ion fluxes, can be impaired by specific antibodies directed against ABP1 and treatment with a C-terminal peptide of ABP1 can mimic infra-optimal concentrations of auxin [111–113]. The null Arabidopsis abp1 mutant is lethal at the embryonic stage and exhibits defects in cell elongation, division location and orientation [9]. Modulations of ABP1 activity in vivo have established that ABP1 is involved in the auxin control of cell expansion and cell division, two of the major cellular responses controlled by auxin and responsible for plant growth and development [114–118].
The next steps in ABP1 characterization will be to identify direct interactors at the PM or in the ER and the components of the transduction chain linking the perception of auxin by ABP1 and downstream auxin responses. We can assume that the two auxin signalling pathways involving the two kinds of auxin receptors, TIR1/AFBs and ABP1, respectively, may interfere with each other in order to integrate the multiple auxin signals and to control plant growth and development with the best accuracy.
5 Conclusion
Auxin action results from the coordinate regulation of its quantity and the sensitivity to the hormone in each cell. Many ways to produce auxin have been evolved by the plant, from various precursors or intermediates and subject to tight regulation. Better understanding of auxin production will be obtained through the elucidation of the localization of each pathway, in which cells and at which developmental stage they are functioning to generate dynamic auxin pools [119]. PAT is also crucial for plant development. Whereas efflux carrier PINs are the primary determinant of the direction of auxin fluxes, influx transporters and MDR-PGPs proteins are proposed to generate auxin sinks and to control the levels in the auxin channels [56]. Even if progress in the PAT field has been considerable, many questions are still open and how exactly auxin regulates the subcellular localization of PINs has still to be determined. Likewise, it is important to better understand the involvement of other hormones in the regulation of PAT [120–122].
The numerous possibilities of interactions between elements of auxin response gene regulation have been mentioned and a better characterization of expression patterns for each member of the ARF, Aux/IAA and TIR1/AFBS families as well as a detailed characterization of their relative affinity of interaction will be essential for a more extensive understanding of their respective roles and contributions to auxin responses according to various cell and tissue types. A specific combination of auxin signalling components might be involved at the resolution of a unique cell or of a small group of cells as recently reported for root organogenesis [38,123]. In addition, the precise contribution of early auxin response genes in the control of cellular responses is still not well understood and little is known about downstream targets.
Our knowledge in auxin responses mediated by ABP1 has improved but the signalling cascade downstream of ABP1 is unknown. Multiple signalling components such as MAP kinases [124,125], IBR5 protein phosphatase [126,127] or RAC GTPases [128,129] have been reported to be involved in auxin signalling but have not yet been implicated in the short auxin signalling SCFTIR1/AFBs pathway. TIR1/AFBs and Aux/IAAs are mainly located in the nucleus, thus physical interaction with ABP1 is highly unlikely. Discovery of how the TIR1 and ABP1 pathways interact is one of the coming challenges.
In this review, the high level of complexity of the regulation of auxin signalling has been discussed and the incredible amount of regulatory inputs requires the development of novel approaches. Modeling of biological processes, assisted by bio-informatic methods, represents a new approach to integrate all existing data and to evaluate the robustness of predictive working hypotheses. The auxin field has already benefited from this approach especially in modeling auxin fluxes just by computing data on PINs localization [130]. We can hope that in the future we will be able to compute all auxin data and to integrate it to plant development as a whole.
Acknowledgements
We are very grateful to Barbara Kunkel for fruitful comments, critical reading and editing of the manuscript. AT had a grant from the French ministère de la recherche et de la technologie.


