1 Introduction
Ventiella sulfuris (Amphipoda, Uristidae) was first described by Barnard and Ingram in 1990 [1] and represents the only-known species of the genus Ventiella. Contributing to more than 98% of the EPR vent amphipods, V. sulfuris is considered as one of the most abundant vent-endemic amphipod species inhabiting the Eastern Pacific Rise (EPR) vents [2,3], but has also been encountered in Manus Basin [4]. This species is frequently observed near colonies of Pompeii worms Alvinella pompejana [5] but also on mussel beds or associated with Riftia pachyptila colonies [6,7]. Even if amphipods have frequently been recorded as a major component of the vent macrofauna, little is known about their ecology or adaptations to vent environments [8,9].
As regards to the extreme conditions prevailing in hydrothermal vent ecosystems, symbiotic interactions between chemoautotrophic bacteria and metazoans appear to be the rule and can be considered as one of the most successful adaptations to these environments [10,11]. Many hydrothermal organisms derive their nutrition from chemoautotrophic bacteria through symbioses relying most often on sulphide or methane as an energy source [12,13]. So, species endemic to hydrothermal vents could be considered as potential hosts for symbiotic interactions with microorganisms. The main objectives of this study were to highlight the occurrence of bacterial symbiosis in V. sulfuris and to hypothesise about their implications in nutrition.
The presence of bacterial symbioses in crustaceans, more often as ectosymbioses has been reported in several vent species such as galatheids Kiwa hirsuta [14] and Kiwa sp. nov [15]; the hermit crab Paragiopagurus ventilatus [16] and in the barnacle Vulcanolepas osheai [17]. The vent shrimp Rimicaris exoculata exhibits the most particular ectosymbiosis in vent crustaceans [18,19]. This shrimp harbours an ectosymbiotic bacterial community (i.e. mainly Gamaproteobacteria, Epsilonproteobacteria) in its expanded gill chamber and on its mouth parts [20–23]. Moreover, a digestive symbiosis involving Deferribacteres, Mollicutes, Epsilonproteobacteria, and Gammaproteobacteria has also been described in R. exoculata's midgut, further suggesting that bacterial community could significantly contribute to the shrimp nutrition [24,25].
In arthropods and more specifically in crustaceans, digestive symbioses involving microorganisms have been studied for decades [26–28] in different groups (i.e. isopods, amphipods and decapods). Bacteria or even fungi can be involved and are often attached to the cuticular lining of the posterior part of the digestive tract (hindgut). Bacteria can, however, be found in the midgut, which is lined by endodermal cells with microvillous brush-border [25].
In V. sulfuris, the main question concerns the occurrence, the location, the aspect and identity (group affiliation) of eventual bacterial symbionts on the integument or in the gut. Microscopic and ultrastructural observations have been focused on the external integument, the gut content and the lining of the different functional parts of the digestive tract. Phylogenetic analyses based on the 16S rRNA genes were used in an attempt to describe the bacterial diversity associated with V. sulfuris.
2 Materials and methods
2.1 Specimen collection
V. sulfuris specimens were collected together with tubeworm colonies of Alvinella Pompejana during the American cruise “LADDER II” (December 2006) at the East Pacific Rise hydrothermal vent site Bio9 (9°50.3′N, 104°17.48′W; 2508 m depth). These colonies were collected by the deep submergence vehicle (DSV) ALVIN operating from the RV “Atlantis”. On board, Alvinella colonies were immediately washed three times with seawater to wash off the sediment and the associated vagile fauna. Washings were sieved on a 500 μm mesh to retain the associated fauna. Immediately after collection, living V. sulfuris specimens were fixed in a 2.5% glutaraldehyde-seawater solution at pH 7.2 and preserved in a seawater-NaN3 6.7 mM solution.
2.2 Microscopy
2.2.1 Light microscopy (LM) and transmission electron microscopy (TEM)
Light and electron microscopic observations were performed on V. sulfuris specimens to locate and identify the occurrence of potential symbiotic bacteria. The external cuticle and gut lining were considered and observed for a total of 15 specimens. Three segments of the digestive tract were considered and observed separately (Fig. 1), the foregut with the stomach (stomodeum), the midgut (mesenteron) and the hindgut (proctodeum).
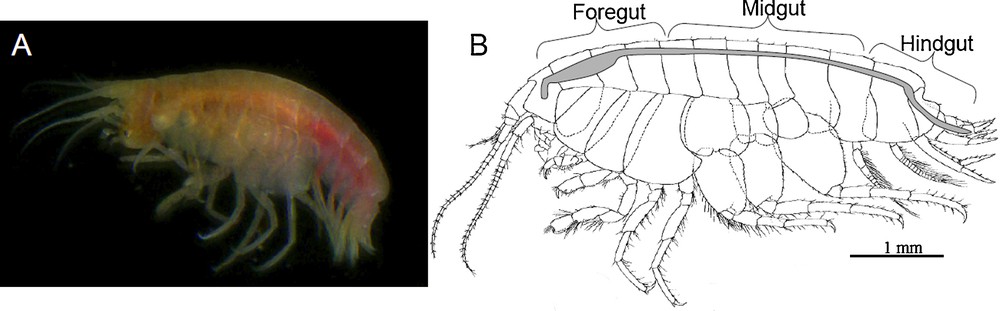
Ventiella sulfuris. General view (A) and morphology of the digestive system (B), which is composed of three main regions (from front to back): the foregut, the midgut and the hindgut.
Entire specimens of V. sulfuris fixed in glutaraldehyde (n = 5) were post-fixed in 1% aqueous OsO4, dehydrated in an ethanol-propylene oxide series and then embedded in epoxy resin (SPI-PON 812, SPI-CHEM). In order to investigate the general organisation of the digestive tract and associated tissues, serial semi-thin sections (1–2 μm) were performed transversally and longitudinally in two specimens with a spacing of 150 μm and 50 μm respectively. The remaining specimens (n = 3) were used to confirm the presence of bacteria at specific levels of the digestive tract. Semi-thin and ultra-thin sections were realized by use of a diamond knife on a Reichert-Jung Ultramicrotome (Ultracut E). Semi-thin sections were then stained with toluidine blue (pH 9.0) for photonic microscopy (Olympus SZ40). Classical uranium acetate and lead citrate stains were applied to contrast ultrathin sections for observation in a Jeol (JEM 100-SX) TEM operating at 80 kV and Tecnai G2 Twin TEM/STEM operating at 200 kV and fitted with an X-ray microanalyser (EDAX).
2.2.2 Scanning electron microscopy (SEM)
SEM observation of the external morphology (two specimens), gut content and gut lining (eight specimens) were realized in a SEM JEOL (JSM-840A) and a ESEM-FEG FEI XL-30 operating at 20 kV accelerating voltage. Fixed-samples were dehydrated through an ethanol series followed by critical point-driying and platinum-coating (20 nm) in a Balzers SCD-030 sputter unit.
2.3 Molecular analyses
2.3.1 16S rRNA gene sequences analyses
Three glutaraldehyde-fixed specimens of V. sulfuris were dedicated to molecular analyses. Due to the small number of specimens and the inadequate fixation, the external carapace of specimens (n = 3) was removed under sterile conditions and discarded to limit contamination by exogenous bacteria. The inner tissues with the gut-associated microorganisms were kept and pooled together. The total DNA was extracted using the FastDNA® SPIN kit for soil (QBIOgen, Santa Ana, CA, USA) following the manufacturer's instructions and stored at 4 °C. Amplifications of the bacterial 16S rRNA genes were performed with universal primers E8F/U1492R (respectively 5′AGAGTTTGATCATGGCTCAG3′ and 5′GTTACCTTGTTACGACTT3′, 1384 bp, annealing temperature 49 °C) or E338F/U1407R (respectively 5′ACTCCTACGGGAGGCAGC3′ and 5′GACGGGCGGTGWGTRCAA3′, 1071 bp, annealing temperature 54 °C). Amplifications were performed on a robocycler GeneAmp™ PCR System 9700 (Applied Biosystems, Forster City, CA, USA) under the following conditions: 3 min at 94 °C, then 30 cycles of 1 min at 94 °C, 1.5 min at the annealing temperature and 2 min at 72 °C, and a final step of 6 min at 72 °C. DNA was amplified in a 50 μL reaction mix composed of 5 μL Taq buffer 10X (QBIOgen), 1 μL dNTP mix 40 mM (QBIOgen), 0.2 μL of each primer 100 μM (Eurogentec, Liège, Belgium), 0.5 μL Taq polymerase 5 U/μL (QBIOgen) and approximately 30 ng DNA template. PCR products were cloned using the TOPO® XL Cloning kit (Invitrogen, Carlsbad, CA, USA) following the manufacturer's instructions. Positive clones were treated for sequencing at the “Plateforme Biogenouest” (Roscoff, France, http://www.sb-roscoff.fr/SG/) with an Abi prism™ 3100 GA using the Big-Dye Terminator V3.1 (Applied Biosystems).
2.3.2 Phylogenetic analyses
Phylogenetic analyses were performed to assess sequence affiliations. Sequences were compared to the Genbank database using the BLAST network service [29]. They were aligned using CLUSTALW and edited using SEAVIEW [30]. Phylogenetic trees were constructed using PHYLO-WIN [30]. The robustness of inferred topologies was tested using 500 bootstraps re-sampling of the trees [31] calculated on the basis of evolutionary distance (neighbour-joining algorithm, [32]) with Kimura two parameters correction matrix. Sequences displaying more than 98% similarity were considered to belong to a single phylotype and were clustered together. Only homologous positions were included in the final alignment. Sequences are available from the EMBL nucleotide sequence database under accession numbers FN429814 to FN429863.
3 Results
The in-depth SEM examination of three specimens of V. sulfuris revealed that their outer cuticular body surface including appendages and gills was devoid of bacterial colonization. Microscopic investigations were then focused on the gut lining and gut content with special attention to the different functional parts of the digestive system (Fig. 1) including the digestive gland. In contrast to the body surface, the digestive tract exhibited two areas of colonization by abundant bacteria that were recurrent in all observed individuals (n = 13) and retrieved in the midgut and the hindgut respectively.
3.1 Morphology and content of the digestive system
3.1.1 General features
Like that of other Amphipods [33,34], the digestive system of V. sulfuris consists of a foregut, a midgut and a hindgut (Fig. 1B). The foregut and the hindgut derive from stomodeal and proctodeal ectoderm, respectively, and appeared cuticle-lined throughout their length. The midgut derives from endoderm and has no cuticle. It appeared as the longest portion of the digestive tract connecting to the digestive gland (two pairs of ventral caeca) and to three dorsal digestive caeca (i.e. a single anterior caecum oriented forward, and a pair of posterior caeca oriented backward).
3.1.2 Digestive content
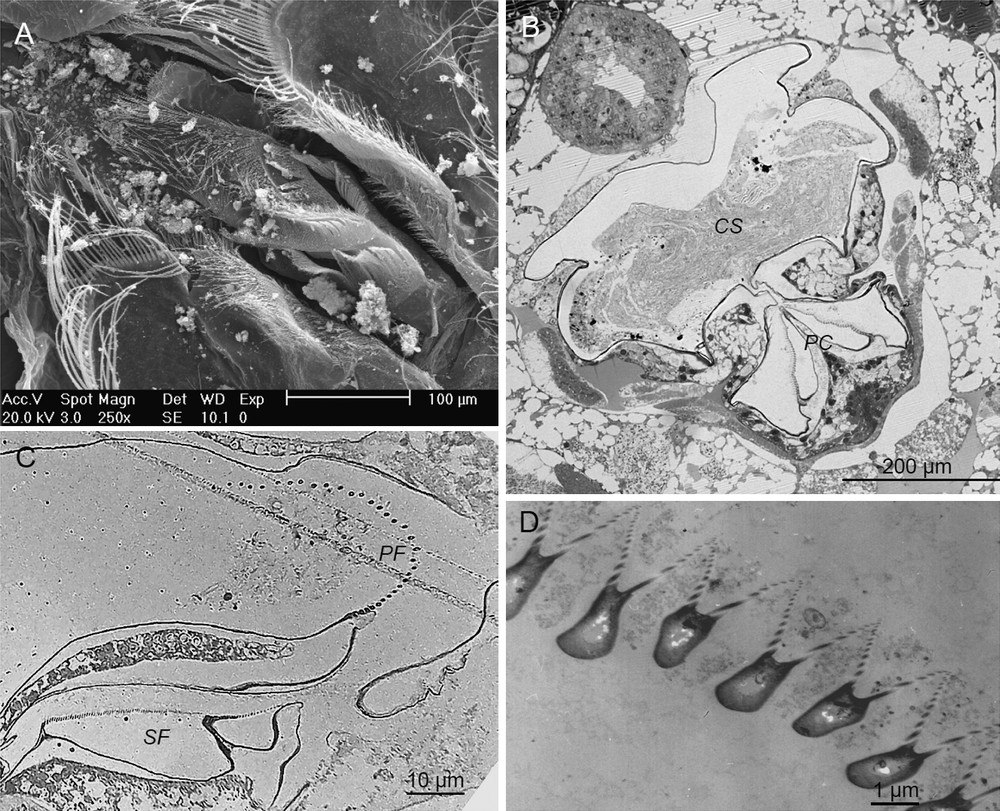
Prevailing in the digestive content of V. sulfuris, large pieces of non-arthropod cuticle were observed all along the digestive tract (Fig. 2A–D). In the stomach, these cuticle fragments exhibited a two-layer architecture with an outer thin electron-dense layer, the epicuticle, and a thick layer constituted by large fibrils of collagen (bundles microfibrils of approximately 100 nm) (Fig. 2B). This architecture of collagen fibrils was easely recognized as an orthogonal plywood arrangement that is typical of the cuticle of annelids [35] and can be identified as belonging to Pompeii worms, Alvinella pompejana [36]. These cuticle fragments appeared more or less altered along the digestive tract (Fig. 2D) then showing separated microfibrils. They also were often accompanied by numerous annelid setae, easily recognizable in cross-section (Fig. 2C) [37] that most probably originate from the alvinellid worms.

Digestive content. A, B. TEM images illustrating the main features of the gut content in the stomach (A, B, E–G) and in the midgut (C, D). A–D. Intact (A, B) and altered (C, D) fragments of alvinellid cuticle (ac) and alvinellid setae (as) with obvious collagen microfibrils (cmf) and fibrils (cf). E. Bacteria (b) and cell membrane debris (mb). F. Bacteria with alvinellid cuticle fragments. G. Intact bacteria, bacterial ghosts (bg) and sheaths (bs).
Besides these abundant fragments of alvinellid cuticle and setae, in the stomach, TEM observations revealed the presence of cell membrane debris (vesicles and folded membranes, Fig. 2E) as well as of numerous bacteria (Fig. 2F) and bacterial traces (empty walls or sheaths) (Fig. 2G) that were associated with the bolus. Microscopic observations of the digestive content occasionally revealed recognizable crustacean cuticle fragments and mineral particles (2–5 μm) identified as metal sulphides (not illustrated).
3.1.3 The foregut
In V. sulfuris, the foregut is lined by a cuticle and consists of oesophagus and stomach. The latter is subdivided into three parts: anterior cardiac region acting in food gringing and mixing, a posterior pyloric filter and a posterior connecting tube or funnel region. The funnel region is a simple tube, devoid of setae. The cardiac stomach is separated from the pyloric chamber by epithelial ridges and linear arrangement of setae (Fig. 3A, B). The pyloric stomach is separated into dorsal and ventral chambers by interlocking setae of the ventro-lateral ridge. In the pyloric region, the structure of the filter chamber (Fig. 3B, C) is typical of amphipods [33]. The secondary filter is composed of parallel rows of comb-like filter bars of 1 μm apart (Fig. 3C). The setae are Y-shaped and bore two rows of setules. The width of the slits between the setules is about 0.06 μm (Fig. 3C-D).
Foregut. A. SEM picture of the stomach. B. Semi-thin cross-sections of both cardiac stomach (Ca) and pyloric chamber (Py) with filter membranes. C. Detailed view of primary filter (pf) and secondary filter (sf) in the pyloric chamber. D. TEM micrograph of the secondary filter showing typical Y-shaped setae.
3.1.4 The midgut and caeca
In amphipods, the midgut or mesenteron and the digestive gland are the major organs involved in digestion and absorption processes of nutrients. Their main common features are the lack of cuticular covering and apical brush-border of their epithelial cells. The midgut is lined by a typical endoderm of cuboidal epithelial cells with short microvilli (approximately 0.5 μm in length). These cells appeared poor in organelles, occupied by a very large nucleus and supported by a thick folded basal lamina. On all observed specimens, electron microscopy observations indicated that the posterior part of the midgut harboured a large community of long, rod-shaped bacteria of about 0.2 μm in diameter and several μm in length (Fig. 4A–E). At high magnification, a thick envelope with a peptide-glycan layer (Fig. 4E) indicated that they are Gram-negative bacteria and the absence of any constriction or separation suggested that they are single-celled. SEM and TEM images also suggested that they preferentially had a longitudinal orientation in the midgut so that most of them were lying on the microvillous brush-border of the midgut epithelium. Longitudinally sectioned bacteria showed that, at the contact with the midgut epithelium, their tip became slimmer than their back filamentous part and curved to be inserted between the microvilli of the healthy cells. Some of the bacteria were in contact with the endodermal cell membrane (Fig. 4C, D) while the filamentous part was located in the ectoperitrophic space (Fig. 4B). In addition, observations at high magnification showed the presence of numerous membrane vesicles and debris between the bacteria (Fig. 4D).
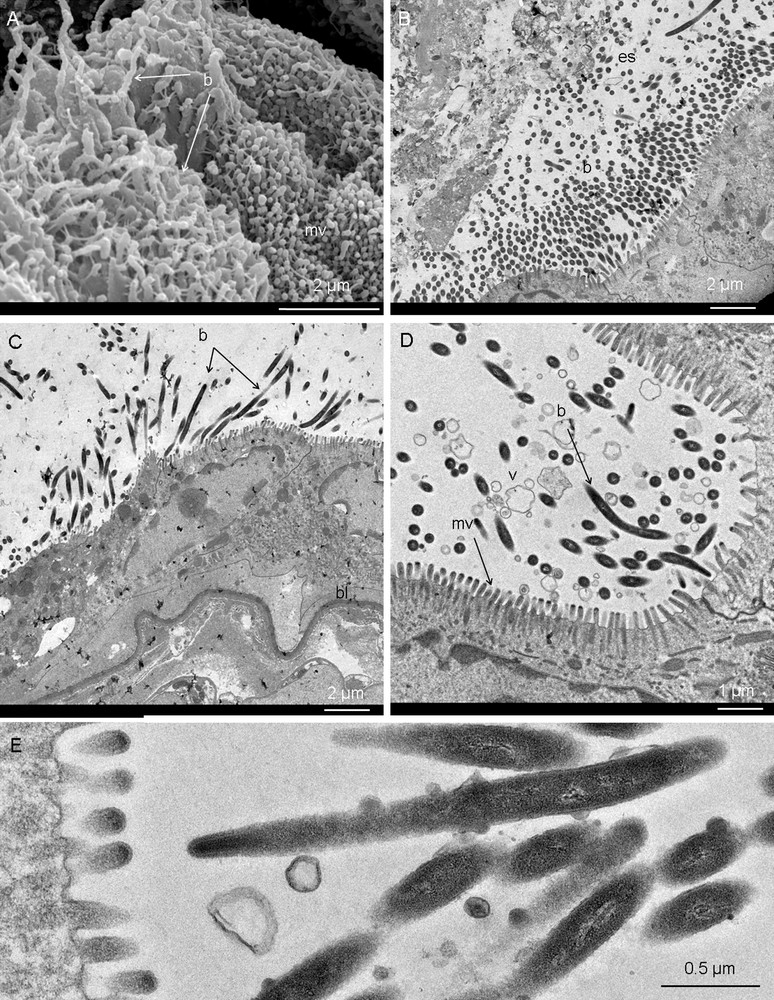
Midgut. A. SEM view of the midgut cells apex showing bacteria (long filaments, b) laying on and between microvilli (mv). B. TEM general view of the midgut epithelium and the high density of bacteria (b) in the ectoperitrophic space (es), i.e. between the gut content and the midgut wall. C–E. Details of the bacteria inserted between the microvilli of the midgut. bl, basal lamina; v, membrane vesicles.
The digestive gland or hepatopancreas consists in four diverticules or caeca that branch ventrally at the junction between the foregut and the midgut. Each diverticule or caecum is lined by columnar epithelial cells bearing apical microvilli (approximately 1 μm in length). In the ventral caeca, the epithelium was differentiated into R/F and B-cells [38] suggesting more intense digestion and absorption activities than in the midgut. Posterior caeca form a pair of midgut extensions rather involved in excretion and reabsorption. The observations of the different digestive caeca of V. sulfuris on semi-thin and ultrathin sections respectively, revealed the absence of bacteria in these structures.
3.1.5 The hindgut
In transverse sections, the hindgut or proctodeum consists in a thick folded epithelium surrounded by muscles (Fig. 5C) in its posterior part (rectum). The hindgut epithelium is lined by a thin cuticle bearing short cuticular projections or spines oriented backward. Some cells exhibited numerous mitochondria that seemed larger than those found in other regions of the digestive system. At the cuticle surface, SEM and TEM observations of V. sulfuris hindgut revealed the presence of densely packed epimural rod-shaped bacteria (Fig. 5A–D). At high magnification, two distinct morphotypes were obvious (Fig. 5D–F): short rods (0.4 × 1 μm) and thin long rods (0.3 × 3 μm) that appeared electron-lucent and electron-dense respectively. Both were tightly attached to the cuticle or cuticular spines (Fig. 5F), formed dense mats in the grooves between the villous folds and appeared embedded in a dense organic matter (Fig. 5E, F).
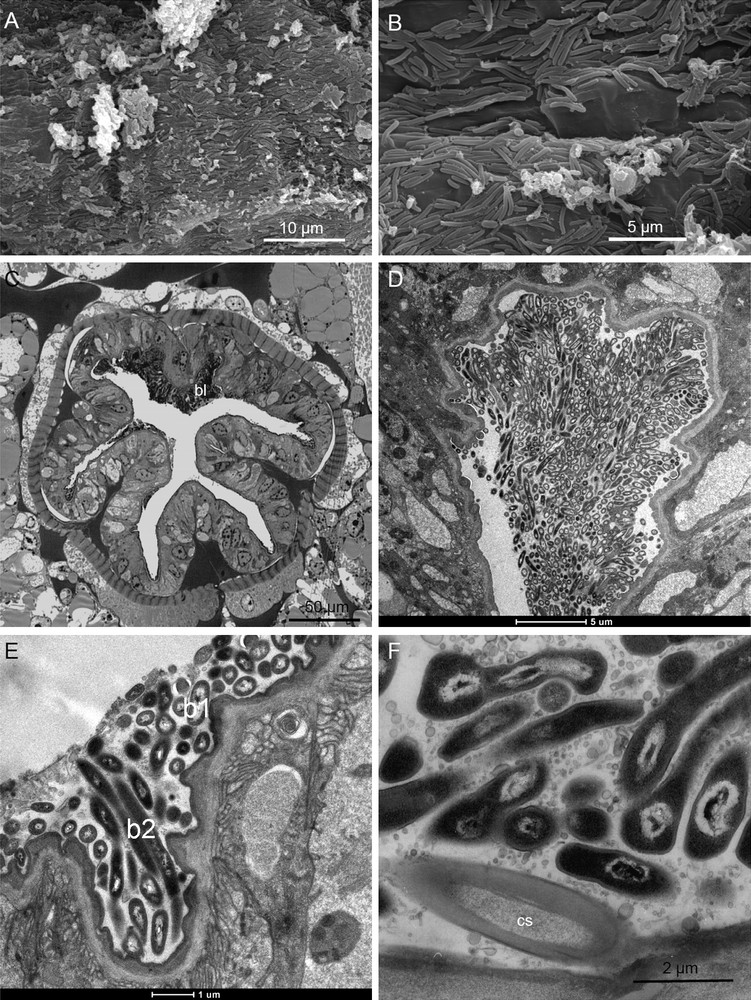
Hindgut. A, B. SEM views of the hindgut wall, massively colonized by rod-shaped bacteria and showing some filaments of few long rods (arrows). C. Semi-thin cross-section in the hindgut exhibiting a dense bacterial layer (b) on its upper side of the rectum. D. TEM view of a dense bacterial mat between villous folds. E. Detail of bacteria embedded in an organic matter and showing two distinct morphotypes, i.e. electron-lucent short rods (b1) and electron-dense long rods (b2). F. Bacterial anchorage on the cuticular spines (cs) of the hindgut. c, cuticle; m, muscle layer; mf, apical membrane infoldings. Masquer
Hindgut. A, B. SEM views of the hindgut wall, massively colonized by rod-shaped bacteria and showing some filaments of few long rods (arrows). C. Semi-thin cross-section in the hindgut exhibiting a dense bacterial layer (b) on its upper side of ... Lire la suite
3.2 Bacterial diversity
Analyses of the bacterial diversity associated with V. sulfuris were done according to the observation of bacteria in the digestive tract. To go further in the description and despite the weak number of remaining specimens, 16S rRNA gene diversity has been analyzed in the amphipod tissues after removal the external cuticle under sterile conditions (three pooled specimens). A total of 103 clone sequences was obtained (65 were amplified using primers E8F/U1492R and 40 using E338F/U1407R) and 97 sequences were analyzed (sequences showing less than 300 bp were excluded). Molecular analysis of the microbial diversity revealed 12 phylogenetic groups among which the phyla predominantly encountered were the Epsilonproteobacteria (21 clone sequences), the Firmicutes (18), the Cytophaga-Flavobacter-Bacteroides (CFB, 14), the Gammaproteobacteria (12), the Betaproteobacteria (12) and the Alphaproteobacteria (6) (Table 1).
Number of bacterial 16S rRNA gene clones from Ventiella sulfuris specimens (n = 3).
| Phylogenetic groups | Number of clones |
| Epsilonproteobacteria | 21 |
| Firmicutes | 18 |
| CFB | 14 |
| Gammaproteobacteria | 12 |
| Betaproteobacteria | 12 |
| Actinobacteria | 7 |
| Alphaproteobacteria | 6 |
| Tenericutes | 3 |
| Acidobacter | 1 |
| Chloroflexi | 1 |
| Deinococcus | 1 |
| Fibrobacter | 1 |
| Total | 97 |
Bacterial sequences from V. sulfuris appeared to be close either to bacteria of other animal guts and most of them clustered with bacteria from arthropod guts or from hydrothermal vents (Table 2, Fig. 6). Indeed, most of the Firmicutes (18) and CFB (14) clone sequences were related to sequences from mammal gut microflora. More interesting, among the six clone sequences affiliated to Alphaproteobacteria, two (i8, e72) were directly related and two appeared close (i32, i71) to bacteria of gut environments of insects or crustaceans such as the chinese mitten crab, Eriocheir sinensis (clone DQ856522, [39]) and the ant Myrmeleon mobilis (clone DQ163946, [40]). In Betaproteobacteria, two groups of nine sequences (i57) and two sequences (i5) could be from Burkholderia sp. bacteria. Indeed, they are sandwiched between two Burkholderia clone sequences (clone AY965240 and clone AY005032) and the closest sequence of the cluster i57 (100% similarities with the hepatopancreatic bacteria of the fresh water isopod Asellus aquaticus, clone 11AY447042) is also virtually identical to Burkholderia sp. isolate N2P5 (U37342; [41]), a free-living soil bacteria [42].
Closest match between representative 16S rRNA gene clone sequences from Ventiella sulfuris and sequences from genbank, based on the BLAST search engine.
| Phylogenetic group | Representative clone sequences | Closest match (accession no.) | Similarity (%) |
| Proteobacteria | |||
| Epsilonproteobacteria | i26 | Rimicaris exoculata gut clone R62LS (FM881772) | 99 |
| Hydrothermal vent gastropod clone SF_C23-G9 (AY531572) | 97 | ||
| e34 | Hydrothermal vent gastropod clone SF_C23-C6_shell (AY531602) | 96 | |
| Gammaproteobacteria | i7 | Frateuria sp. DM-HM (DQ419968) | 99 |
| Rhodanobacter thiooxydans (AB286179) | 98 | ||
| i28 | Psychrobacter sp. P11-B-2 (EU016144) | 99 | |
| Betaproteobacteria | i5 | Iron-reducing bacterium clone HN4 (FJ269046) | 99 |
| i54 | Pelomonas saccharophila (AM501432) | 99 | |
| i57 | Burkholderia symbiont Asellus aquaticus clone 11 (AY447042) | 99 | |
| Alphaproteobacteria | i8 | Eriocheir sinensis gut clone C2Q (DQ856522) | 94 |
| i32 | Bacterium clone 015C-B05 (AY662021) | 92 | |
| i71 | Bacterium clone Gsoil 264 (AB245345) | 99 | |
| e13 | Hyphomicrobiaceae clone Amb_16S_929 (EF018645) | 97 | |
| e72 | Myrmeleon mobilis (clone DQ163946 ) | 99 | |
| Firmicutes | i64 | Alkalibacterium putridalgicola (AB294170) | 100 |
| e17 | Ovis ammon gut clone AS2_aao35b10 (EU465772) | 97 | |
| e36 | Bovine rumen clone 1103200832524 (EU845714) | 99 | |
| e45 | Mouse cecum clone 16saw39-1f01.w2k (EF604607) | 96 | |
| e60 | Rumen bacterium clone GRC56 (DQ673521) | 96 | |
| e75 | Potamochoerus porcus gut clone RRH_aaa01c03 (EU474931) | 95 | |
| e79 | Tidal sediment clone TfC20H82 (EU362236) | 99 | |
| CFB | e35 | Sediment clone ORSATC_h06 (EF393146) | 99 |
| e61 | Equus asinus gut clone WA_aaa03g03 (EU779398) | 91 | |
| e82 | Equus grevyi gut clone GZ_aaa03a09 (EU470441) | 93 | |
| e84 | Elephas maximus gut clone AE1_aaa04c09 (EU471567) | 93 | |
| e86 | Equus equus gut clone horsem_aai95a09 (EU463500) | 97 | |
| e95 | Bacterium clone 060C09_B_SD_P93 (CR933312) | 92 | |
| Actinobacteria | e27 | Gordonia sp. D2 (DQ787430) | 97% |
| e59, e88 | Knoellia subterranea strain HKI 0120 (AJ294413) | 97–98 | |
| Tenericutes | i70 | Kiwa hirsuta epibiont clone F8 (EU265798) | 96 |
| e43 | Bovine rumen clone 1103200843512 (EU844544) | 99 | |
| Acidobacter | e23r | Sludge bacterium clone S6 (AF234751) | 95 |
| Chloroflexi | e99r | Pachnoda ephippiata midgut clone PeM11 (AJ576402) | 96 |
| Deinococcus | e51r | Denitrifying consortium bacterium clone OTU_23 (EU083501) | 96 |
| Fibrobacter | e92 | Equus grevyi gut clone GZ_aaa01b11 (EU470410) | 94 |
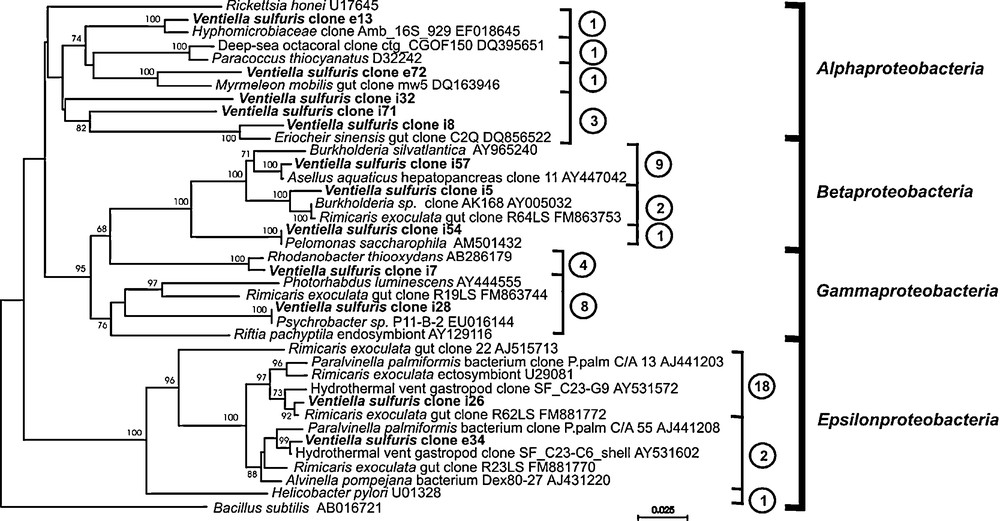
Phylogenetic tree based on proteobacterial 16S rRNA gene sequences from Ventiella sulfuris bacterial clones from EPR hydrothermal fields. The number of clone sequences affiliated to each cluster is in circles.
In contrast to the previously mentioned proteobacterial groups, the clustering of Gamma- and Epsilonproteobacteria-affiliated sequences seemed to be related to deep-sea and/or hydrothermal vent bacteria, and especially to symbiotic bacteria associated with vent organisms. Indeed, the two thirds of the Gammaproteobacteria-affiliated sequences (eight on 12 clones) clustered together in one group (i28) and are very close to the sequence of Psychrobacter sp. (P11-B-2 EU016144), a manganese bacteria from Arctic deep-sea sediment also close to some gut-associated bacteria of the vent shrimp Rimicaris exoculata from the Mid-Atlantic Ridge (MAR, [25]). The other 4 clone sequences are very close to that of a hydrothermal free-living sulpho-oxidizing bacteria (Rhodanobacter thiooxydans AB286179, [43]). The sequences affiliated to Epsilonproteobacteria clustered with clones retrieved in hydrothermal invertebrates such as shrimps, gastropods or worms (MAR, Central Indian Ridge or CIR and EPR) (Table 2, Fig. 6). The great majority of the sequences (18 out of 21) clustered first in one group (i26). Their closest sequence is that of gut bacteria of the vent shrimp R. exoculata (gut clone R62LS FM881772; 92% of similarities). The three other clone sequences obtained in V. sulfuris (e34) clustered to epibiotic bacteria from a vent gastropod (AY531602, [44]) and was also very close to bacteria found alvinellid worms as Paralvinella palmiformis (AJ441208 [45]) and Alvinella pompejana (AJ431220 [46]).
4 Discussion
In this study, a combination of microscopy and 16S rRNA gene sequence analyses were performed to give a first description of a dual bacterial symbiosis in the digestive tract of the vent amphipod V. sulfuris. These two distinctive bacterial colonizations located in the midgut and hindgut respectively, can be considered as typical of symbiotic associations in regard to the morphology (single morphotypes), location, arrangement and recurrence of the bacteria observed on all specimens. Moreover, digestive content analysis suggests that V. sulfuris feed on the A. pompejana colonies, i.e. on the worm epibiont bacteria and/or on the worm tissues. Sequencing analyses of 16S rRNA genes revealed that the bacteria found in the digestive system of V. sulfuris belong to six phyla, three major ones (Epsilonproteobacteria, Firmicutes and Cytophaga–Flavobacter–Bacteroides) and three minor ones (Gammaproteobacteria, Betaproteobacteria and Alphaproteobacteria) in number of obtained clone sequences. Most of the clone sequences are close to symbiotic bacteria of hydrothermal vent organisms and to bacteria involved in digestive symbioses especially in crustaceans. Both microscopy data and 16S rRNA gene sequences analyses let us to categorize V. sulfuris gut bacteria in four groups according to their origin (see [27,47] for review): resident symbiotic bacteria (1), transitory digestive bacteria (2) and ingested bacteria with food (3) or from the environment (4).
4.1 Diet and trophic interaction
V. sulfuris are frequently encountered in different vent biotopes [5–7], co-occurring with colonies of Pompeii worms and other organisms [4,6]. In situ observations of V. sulfuris specimens have however revealed that they have been frequently observed on the dorsal integument of A. pompejana specimens [5]. An hypothesis has been suggested that the amphipod could graze directly on epibiotic bacteria growing on dorsal side of the Pompeii worm body [2,5]. Supporting this hypothesis, evidence that V. sulfuris feed on A. pompejana worms or in their tubes is given by the abundance of A. pompejana cuticle fragments and setae in their gut content. In the same way, several clone sequences (Epsilonproteobacteria) recovered in the gut of V. sulfuris clustered with sequences from alvinellid epibionts, strongly suggesting that the latter were ingested by the amphipod (see below). These first observations confirm that V. sulfuris could ingest epibiotic bacteria of Pompeii worms together with worm tissues, probably from the dorsal expansions. The alteration or disappearance of the bacteria and worm tissue items along the digestive tract also suggested that the amphipod feeds on these items and assimilates their components. The results can however not decide about the real diet of the V. sulfuris species and a feeding strategy on dead A. pompejana individuals (i.e. necrophagous) could not be excluded but the occurrence of fragments of non-degraded cuticle (i.e. intact collagen fibers) rather supports the “grazing” hypothesis. The “necrophagous” hypothesis appeared not likely because the proportion of cell debris in the amphipod gut is low compared to that of cuticle fragments and no traces of inner tissues (e.g. cell nucleus, storage granules, Fig. 2E-G) were observed even in the stomach. Most of the amphipod gut content seemed to come from the worm surface tissues. The cell membrane debris most probably came from the cells extensions protruding through the cuticle [36]. V. sulfuris could then be regarded as a “grazer” on tube worms in different vent biotopes [5–7]. In these cases, its diet seems to be rather bacterivorous/carnivorous but requires confirmation by stable isotope analysis (13C/12C and 15N/14N ratios) to determine the contribution of the various food sources.
4.2 Digestive bacterial symbioses
The significance of symbiotic relationships in deep-sea reducing environments was not broadly recognized until the discovery of deep-sea hydrothermal vent ecosystems [11,13]. There is now accumulating evidence that microbial colonization of the gut of marine invertebrates is widespread and that these interactions could be mutually beneficial in terms of nutrition [47,48].
Ultrastructural observations of the digestive system of V. sulfuris revealed at least two symbiotic communities of epimural resident bacteria, in the midgut and in the hindgut respectively. Both communities fulfil the accepted morphological criteria [27,49–51] of bacterial symbioses possibly mutualistic and/or obligate, i.e.: (i) the recurrence of the association and bacterial cell morphotypes in all specimens; (ii) the recurrence of the locations of bacterial settlements, and (iii) the healthy appearance of the tissues in contact with the bacteria.
The molecular identification of the gut bacteria by 16S rRNA genes analyses would also support the presence of bacterial symbiotic associations in the digestive tract of V. sulfuris. Unfortunately, they were rather limited because of a limited number of specimens and of a unsuitable fixation of specimens (glutaraldehyde 2.5%) that has partly degraded bacterial DNA and did not allow FISH that would be more efficient to correlate bacteria phylogenetic affiliation to their location. Nevertheless, distinct phylogenetic groups of bacteria have been identified and come undeniably from the gut or its gut content owing that the outer integument tissues were removed under sterile conditions. According their closest phylogenetic affiliation, the origin (i.e. environment, ingested food, transitory and resident microflora), location and potential role of some bacterial phylotypes were hypothesized in concordance with the microscopic observations.
Evidence of environmental origin for all the Gamma- and Betaproteobacteria clones as well as for two of the Alphaproteobacteria clone clusters is given by their closest affiliation to free-living bacteria from deep-sea or marine sediment. This is clearly the case for the Betaproteobacteria clone clusters (i5, i57, Table 1) that are close to free-living Burkholderia bacteria. In V. sulfuris, Betaproteobacteria should be regarded at best as members of a transitory microflora. Similarly, few Epsilonproteobacteria (clone cluster e34; Fig. 6) could probably be alvinellid-associated bacteria ingested as/or with food, owing that many bacteria cells and ghosts observed in the bolus (discussed above). They are closely related to bacteria from the scaly-foot gastropod from CIR hydrothermal fields [44] and thus could be rather ubiquitous epibiotic bacteria of vent invertebrates. In contrast, the clone cluster i26 is related to a gut clone of Rimicaris exoculata [25] and could most likely be a midgut-associated bacteria (see below).
In the midgut, the long rod-shaped bacteria inserted between microvilli of endodermal cells could be considered as a long-term resident community because the midgut is the only part of the digestive tract not prone to moulting. Moreover, they showed a very unusual settlement in an absorptive part of the digestive tract. Similar settlement is known for stalk-forming Rickettsiales ‘Candidatus Hepatincola porcellionum’ colonizing the midgut glands of terrestrial isopods [52,53]. The midgut bacteria of V. sulfuris ultrastructurally differ by their envelope (Gram-) and could rather be Epsilonproteobacteria. A very similar morphotype of bacteria was previously retrieved with identical settlement features in the midgut of the hydrothermal vent shrimp, R. exoculata [25]. In the same paper, these authors identified R. exoculata midgut clones as affiliated to Epsilonproteobacteria among which the clone R62LS FM881772 appeared as the closest one to the clone cluster i26 obtained from V. sulfuris. In the same way, such long rod-shaped bacteria have been observed by SEM and TEM in the midgut of several thalassinid shrimps ([54], Compère pers. com.) and Epsilonproteobacteria have also been identified by molecular analyses in one species [55]. Some authors (see [27] for review) interpreted the presence of bacteria in the midgut of crustaceans as an opportunistic or parasitic presence. In terms of attached parasites, the midgut is clearly the place with the biggest benefits [48]. Because of the healthy nature of the midgut epithelium observed in V. sulfuris, we could not adhere to this idea. In contrast, Epsilonproteobacteria are regularly involved in epibiotic, symbiotic or mutualistic interactions with hydrothermal invertebrates [11,13] and most bacteria affiliated to the phyla harbour varied chemoautotrophic metabolisms such as iron-oxidation, sulphide-oxidation or methylated compounds-oxidation in diverse ecosystems. 16S rRNA gene sequences analyses does not permit to infer a role for Epsilonproteobacteria associated with the amphipod gut but suggests that V. sulfuris clones which clustered in this group, could be specific to hydrothermal vents and considered as invertebrate-associated bacteria. Likely located in the midgut, this bacterial population may be rather involved in detoxification or nutritive relationships with its host, as proposed for midgut symbionts of vent shrimp, R. exoculata [25]. In the gill chamber, the ectosymbiosis of shrimp R. exoculata [22,56], such relationships between epsilonproteobacteria and their host are recognized as the rule.
In arthropods and more precisely in crustaceans, the privileged area for bacterial settlement is however the posterior part of the digestive tract, the hindgut, covered by a chitinous cuticle that provides anchoring surfaces for bacteria and favours symbiotic interactions [26,28]. Hindgut colonization by resident microorganisms was especially reported in detritivorous species that ingest recalcitrant, i.e. “hard-to-digest” compounds as certain polysaccharides (cellulose, chitin) and stabilized matters (lignin, sclerotized proteins…) for whose the species do not dispose of appropriate enzymes. The symbiotic bacteria are thus supposed to help in the digestion of the recalcitrant compounds but also to supply the host in nutriments or in organic nitrogen. Among insects, such associations are thoroughly studied notably in wood-eating termites [57–59], coleopteran larvae [60], cockroaches [61] and the cricket Gryllotalpa orientalis [62]. In crustaceans, broad bacterial colonizations of the hindgut walls were described in mud shrimps (Thalassinids) and squad lobsters (Galatheids) from the littoral [49,50,54] and from deep-sea wood falls [51]. Permeability is probably facilitated at this site, which seems to be best developed in termites, cockroaches and coleopterans, in which the uptake of small organic molecules (e.g. short-chain fatty acids, amino-acids) from the hindgut is of nutritional significance [60,63]. It is probable that the hindgut of V. sulfuris also acts in the exchange or absorption of small molecules because its ultrastructural features (thin cuticle, cell apical membrane infoldings, mitochondriae) are characteristic of permeable integument [64] and resemble to those of the hindgut wall in termites [57–59] and coleopteran larvae [60].
According to our observations, symbiotic relations could thus occur in the hindgut of V. sulfuris that appeared to harbour two morphotypes of resident bacteria. The latter could then be regarded as symbiotic bacteria with digestive implications. By deduction and according to the previous considerations, the hindgut bacteria of V. sulfuris are likely among the Alphaproteobacteria, the Firmicutes and/or the CFB identified by the 16S rRNA gene analyses. The Alphaproteobacteria clone sequences (i8, i32, i71, e7) from V. sulfuris are close to symbiotic gut bacteria of the ant Myrmeleon mobilis (clone DQ163946, [40]) and of the Chinese mitten crab, Eriocheir sinensis (clone DQ856522, [39]). The Firmicutes and CFB that dominates in number of clone sequences in V. sulfuris, often contribute to vertebrate and invertebrate gut microflora, as pathogens or commensals [65,66]. They are also retrieved in hydrothermal environments [67]. Some bacteria affiliated to Firmicutes and CFB are known to degrade organic matter, and particularly recalcitrant polymers such as chitin and collagen [68]. In V. sulfuris, the Alvinella cuticle fragments could be considered as a recalcitrant material of which hydrolysis probably requires direct or indirect bacterial action, notably through enzyme synthesis. The prevalence and persistence of alvinellid cuticle fragments in the gut of V. sulfuris may also be explained by the exceptional resistance of their collagen fibres [36,69] that differs from the worm interstitial tissue collagen and from fibrillar collagen of vertebrates by its composition, size, domain structures and immunological properties. Among the fibrillar collagens of 40 other invertebrates and vertebrates, the cuticular collagen produced by A. pompejana is positioned at the upper limit for melting temperature, just before that of thermostable synthetic collagens [69]. It could then be considered as recalcitrant to digestion by organisms and its degradation probably requires specific enzymes. Collagenolytic or proteolytic enzyme activities have already been evidenced in the bone-eating worms of the genus Osedax [70]. These activities are restricted to the root tissues that house symbiotic bacteria, belonging to the Gammaproteobacteria lineage (order Oceanospirillales). Even if direct correlations have not been yet established between symbiotic bacteria and collagenase production, authors suggest that collagenase might help Osedax to exploit organic carbon complexes from the external environment. So, gut bacteria associated with V. sulfuris and affiliated to Firmicutes and CFB phyla could be involved in the degradation of collagen particles from Alvinella cuticles and could be located in the hindgut.
5 Conclusions
Investigated for the first time, the biology of the amphipod V. sulfuris highlighted the close trophic relationships between this vent species and the Pompeii worm A. pompejana in EPR area. Microscopic investigations of the gut of V. sulfuris brought new insights on the diet and importance of bacteria in the digestive system of hydrothermal vent animals. The presence of A. pompejana cuticle fragments and bacterial epibionts in the amphipod gut content suggests that V. sulfuris could directly feed on the worm and notably grazes on the bacterial community colonizing its dorsal integument. Combining morphological observations and molecular analyses on several specimens, different resident bacterial populations have been identified for the first time in the midgut and in the hindgut. The resident bacterial population in the midgut, associated with healthy tissues, could be affiliated to Epsilonproteobacteria cluster, specific to reducing environments and usually involved in symbiotic interactions. As related clones from EPR and MAR invertebrate-associated epibionts, this community could be specific to the amphipod and involved in nutritive or detoxification relationships with the host. According to the clustering of the Alphaproteobacteria clones and to the background of Firmicutes and CFB phyla, this bacterial population could form the resident (and/or transitory) symbiotic microflora in the hindgut of V. sulfuris and could be notably involved in the hydrolysis of recalcitrant organic matter retrieved in the digestive content (e.g. cuticle fragments of Alvinella). At present time, it is, however, not possible to conclude on the putative role and mutualistic implications of these bacterial symbionts without additional investigations.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgments
The authors thank the Belgian Fund for Joint Basic Research (FRFC-Belgium, conv. no 2.4594.07.F) for financial support and post-doctoral research grant of Dr L. Corbari at the University of Liege (Belgium). They also thank the partnership “Hubert Curien Tournesol 2009-2010” for facilitating the exchanges between ULg and Ifremer. The LADDER project was funded by NSF Ocean Sciences grant OCE-0424953. Special thanks to the chief scientists, J. Ledwell and S. Mills and also to the Alvin Team (Woods Hole Oceanographic Institution). The authors would also thank F. Pradillon (Jamstec, Japan) for her precious on-board help during the specimen sampling. Thanks to “Plateforme Biogenouest” for sequencing work and ANR DEEPOASES for financial support. The authors also wish to express their appreciation to N. Decloux (ULg, Belgium) for her excellent technical assistance in TEM and SEM. They also thank the Centre of Applied Technology in Microscopy (ULg, Belgium) for providing access to electron microscopy equipments.


