1 Introduction
A proportion of Indo-Pacific reef fish species exhibit some phylogeographic discontinuity coinciding with the Indo-Pacific barrier ([1,2] and references therein). It has been hypothesized that a majority of these phylogeographically-structured species actually consist of pairs of geminate species, each restricted to a single ocean [3]. This hypothesis, if confirmed, would considerably affect our current understanding of what determines species richness in the Coral Triangle [4]. This would imply shifting the current paradigm of the Coral Triangle as biodiversity epicentre [4] towards the hypothesis that the higher species richness in the Coral Triangle artefactually results, at least in part, from the overlap in the geographic distributions of Indian and Pacific species [2,3,5].
Under the current taxonomic standards, evidence of morphological differentiation is required to support the recognition of separate species. As noted by J.E. Randall [6] about species differentiation between the Indian and the Pacific Oceans: “Many examples of such specific pairs are known […] but perhaps there are others that may have diverged ecologically, behaviourly, or physiologically, but not enough morphologically for us to readily detect”. Molecular population genetics should be considered in the latter, as it offers the tools and concepts to address the problem of cryptic species [7]. Actually, new species discoveries in fishes increasingly stem from molecular genetic studies (e.g. [8–14]).
Here, we examine the taxonomic status of a widespread Indo-West Pacific reef fish, the humbug damselfish, Dascyllus aruanus. The humbug damselfish is common in shallow reef habitats throughout the Indo-West Pacific. It has been used as a model-species for a number of ecological, behavioural, and population genetic studies [15–26]. The phylogeographic structure of the humbug damselfish indicates two main metapopulations across the Indo-West Pacific, each one specific to an ocean [26]. No mitochondrial haplotype characteristic of Indian Ocean individuals has been found in the western Pacific and no Pacific haplotype has been found in the Indian Ocean [25,26]. The phylogeographic break as determined from mitochondrial haplotypes is located at the eastern edge of the Sunda Shelf [25]. Nuclear markers (microsatellites) have also been used for a similar distribution-wide analysis of geographic structure in humbug damselfish [26], showing a same distinct genetic break between the Indian and Pacific Oceans that could not be explained by mere isolation by distance [26]. These observations have been interpreted as the result of vicariance on either side of the Indo-Pacific barrier [26]. It has been hypothesized that reproductive barriers currently limit gene flow between humbug damselfish populations from the Indian Ocean and the Pacific Ocean [25].
We review the phylogeographic information available on the humbug damselfish and provide the geographical delineation of the two main mitochondrial lineages previously uncovered in this species [3,25–27]. The combined evidence from both mitochondrial DNA and nuclear DNA, and pigmentation patterns allows us to address the issue of cryptic speciation in the humbug damselfish.
2 Materials and methods
2.1 Review of published sequence data
Previous studies have produced the mitochondrial sequences of humbug damselfish from a total of 43 sampling sites throughout the Indo-West Pacific [3,20,25–27] (Fig. 1A). The published mitochondrial data selected for the present survey thus included the complete ATP6/8 gene sequences [842 base pairs (bp)] of 18 individuals from 10 sampling sites in the Indo-West Pacific [27], the partial cytochrome-oxidase I gene sequences (842 bp) of 11 individuals from two sampling sites in the southwestern Indian Ocean and one sampling site in the central Pacific [3], the partial control-region sequences (approximately 359 bp) of 401 individuals from 18 sampling sites in the Coral Triangle [25], and the partial cytochrome b gene sequences (1058 bp) of 260 individuals from 13 sampling sites throughout the Indo-West Pacific [26] (Supplementary material, Table S1). In each of the foregoing surveys, haplotypes have been determined as belonging to either one of two haplogroups, here coined “Indian” and “Pacific”. The humbug damselfish population of Cebu Strait, Philippines was sampled by both J.-M. Raynal et al. [25] and S.-Y.V. Liu et al. [26]. The haplotype composition of the Cebu sample in [26] was therefore used to establish haplogroup relationships of the control-region haplotypes produced by [25]. The partial control-region sequence from an individual from Glorieuses Islands, southwestern Indian Ocean (C. Fauvelot, unpublished) clustered with haplotypes of the Central Java Sea, thus establishing that the latter belonged to the “Indian” mitochondrial type (mitotype) [25]. In addition, we examined the set of partial control-region sequences (387 bp) of 45 individuals from 5 sampling sites in French Polynesia [20]. These sequences, accessible from Genbank (http://www.ncbi.nlm.nih.gov/; accession Nos. KM406787–KM406831) were included in a single FASTA file together with reference control-region sequences of the Indian and Pacific mitotypes, and the whole matrix was subjected to Neighbour-Joining analysis using the Mega 5 package [28].
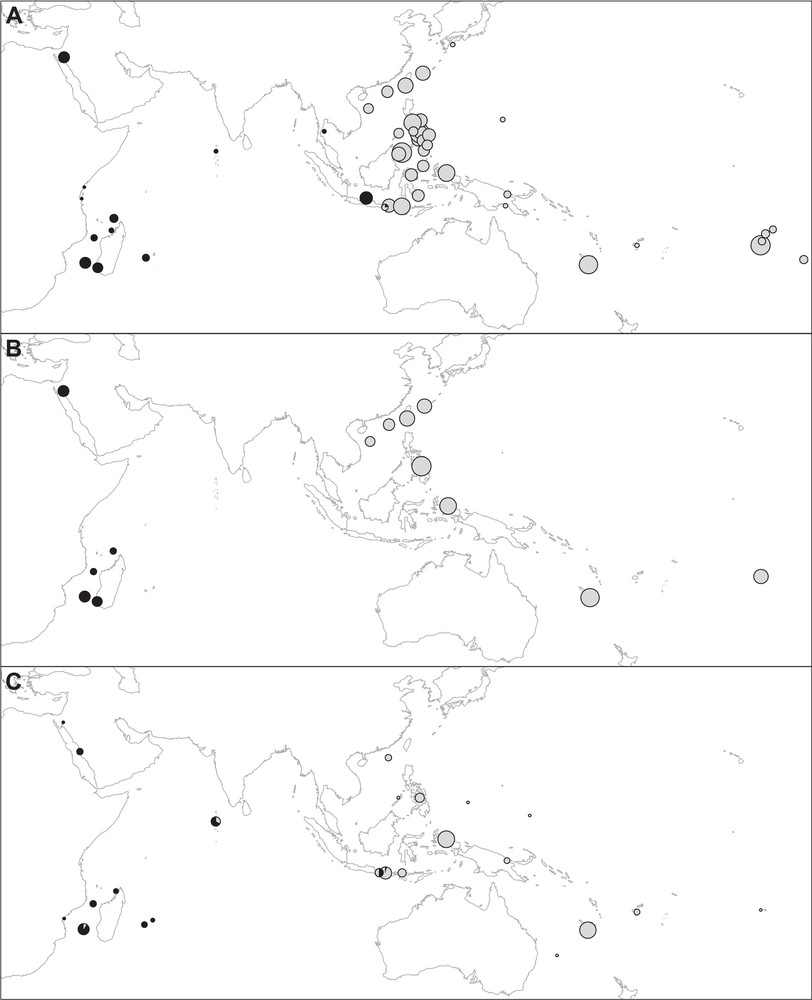
Summary of phylogeographic data on humbug damselfish (Dascyllus abudafur and D. aruanus). Circle area proportional to sample size. A. Geographic distribution of mitochondrial types from a compilation of published and unpublished sources ([3,20,25–27]; present study). Black, Indian Ocean mitotype characterizing D. abudafur; light-grey, Pacific Ocean mitotype characterizing D. aruanus. B. Geographic distribution of nuclear types inferred from microsatellite genotypes [26]. Black, genotypes assigned to D. abudafur; grey, genotypes assigned to D. aruanus. C. Pie diagrams representing the frequencies of individuals with black or solid-grey (black), and light-grey or white (light-grey) basis of the caudal fin.
Similarly, multi-locus microsatellite genotypes from 13 samples from sites across the Indo-West Pacific (Fig. 1B) were determined to belong to either an “Indian” or a “Pacific” form according to their probability score under Bayesian structure analysis [26].
2.2 Additional sampling and sequencing
The cytochrome b gene sequences of an additional sample of 6 individuals from Madang, Bismarck Sea were produced according to the methods and protocols detailed in [26]. Another 5 individuals from Serangan, on Bali Island, western shore of Lombok Strait were also sequenced at the cytochrome b locus, using primers HIMB01 (5′-GTGACTTGAAAAACCACCGTTG-3′) and HIMB02 (5′-AATAGGAAGTATCATTCGGGTTTGAT-3′). Polymerase chain reaction was done in 25 μL volumes, using 1 μL of template DNA, 4 μL 10 × PCR buffer (Applied Biosystems, Foster City CA, USA), 2.5 μL 10 mM dNTPs, 1.25 μL of each primer at 10 mM, 2 μL 25 mM MgCl2 solution, 0.125 μL AmplyTaq Gold™ (Applied Biosystems) in double-distilled H2O. The thermocycling profile included an initial denaturation of 94 °C for 15 s, 38 cycles of 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 45 s, with a final extension of 72 °C for 5 min. All 11 new sequences were then determined to haplogroup. For this, we compared them to previously published cytochrome b gene sequences [26] by checking their nucleotide composition at diagnostic nucleotide sites under Mega 5 [28]. Control-region sequences of another batch composed by 1 individual from Zanzibar, 3 individuals from the Mozambique channel, 5 from the Philippines, 1 from Guam, and 2 from Fiji, produced according to the protocols exposed in [20], and using primers CR-A-DA (5′-ATGAATCTTACAACTCAACACCTG-3′) and CR-E-DA (5′-TCAACCAAGTACAACCCCTGT-3′) specifically designed by one of us (CF) for D. aruanus were added, and similarly determined to one of the two main haplogroups.
2.3 Pigmentation patterns
Specimens from throughout the distribution of the species were photographed either live underwater or after being captured. The photographs were subsequently examined for pigmentation patterns. Additional photographs were retrieved from FishBase (http://www.fishbase.org/; [29]), and from the Barcoding of Life database (BOLD) (http://www.barcodinglife.com/). The sampling sites for individuals sampled for pigmentation patterns are reported in Fig. 1C. We focused on the intensity of the pigmentation of the blackish blotch at the basis of the caudal fin as this was found to be variable among individuals. Four patterns were determined, from blackish (Fig. 2A; scored 3) to lacking pigmentation (Fig. 2B; scored 0) with two intermediate patterns, where the basis of the caudal fin appeared light-grey (scored 1) or solid-grey (scored 2). The samples examined for pigmentation patterns were from the Red Sea (n = 6 individuals), the Mozambique Channel (n = 24), the Mascarene Islands (n = 6), the Maldives (n = 10), Bali (n = 8), Lombok (n = 18), Komodo (n = 8), the South China Sea (n = 5), the Philippines (n = 11), Raja Ampat (n = 35), the Bismarck Sea (n = 4), Chuuk (n = 1), Yap (n = 1), Lord Howe (n = 1), New Caledonia (n = 32), Fiji (n = 4), and the Society Islands (n = 1) (details in Supplementary material, Table S2 and Figs. S1–S7). Mitochondrial sequences were available for 55% of the Indian Ocean individuals and 41% of the Pacific Ocean individuals examined from photographs. “Indian Ocean” here refers to the region of the Indo-Pacific west of Lombok Strait, while “Pacific Ocean” refers to the region north and east of Lombok Strait.

Photographs that illustrate the differences in pigmentation patterns between Dascyllus abudafur and D. aruanus. A. D. abudafur 64 mm standard length (SL) from Juan de Nova Island, Mozambique Channel; pigmentation score of the blackish blotch at the basis of the caudal fin is 3. B. D. aruanus 45 mm SL from Raja Ampat, West Papua; pigmentation score is 0.
2.4 Statistical analyses
The Mantel test implemented in Genetix 4.02 [30] was used to test the correlation between pairwise θ estimates at the 7 microsatellite loci scored by [26] and their homologues at the cytochrome b locus [26]. Net nucleotide distance between the Indian and Pacific mitochondrial haplogroups, based on Kimura-2 parameter nucleotide substitution model, was estimated under Mega 5 [28]. A χ2 test of homogeneity was used to test the null hypothesis of no morphological difference between Indian and Pacific forms, as defined by, respectively, geography and mitotype.
3 Results
The frequencies of Indian and Pacific mitochondrial haplogroups per sampling site are given in Supplementary material, Table S1 and reported on Fig. 1A. New cytochrome b and control-region sequences are presented in Fig. 3 and in Supplementary material, Tables S3–S5. The new cytochrome b gene sequences from Lombok Strait (n = 5) comprised one sequence clustering with the previously identified Indian haplogroup, the remainder being of the previously identified Pacific haplogroup. Those from the Madang sample (n = 6) were all found to belong to the Pacific haplogroup. The control-region haplotypes sampled in Zanzibar and Glorieuses Islands all belonged to the Indian Ocean haplogroup whereas all control-region haplotypes from the Philippines, Guam, Fiji, and French Polynesia belonged to the Pacific Ocean haplogroup, as confirmed by Neighbour-Joining analysis. The nucleotide distance between the Indian and Pacific mitochondrial haplogroups, estimated from cytochrome b gene sequences, was 0.6% (Mega 5; Kimura-2 parameter net distance [28]).
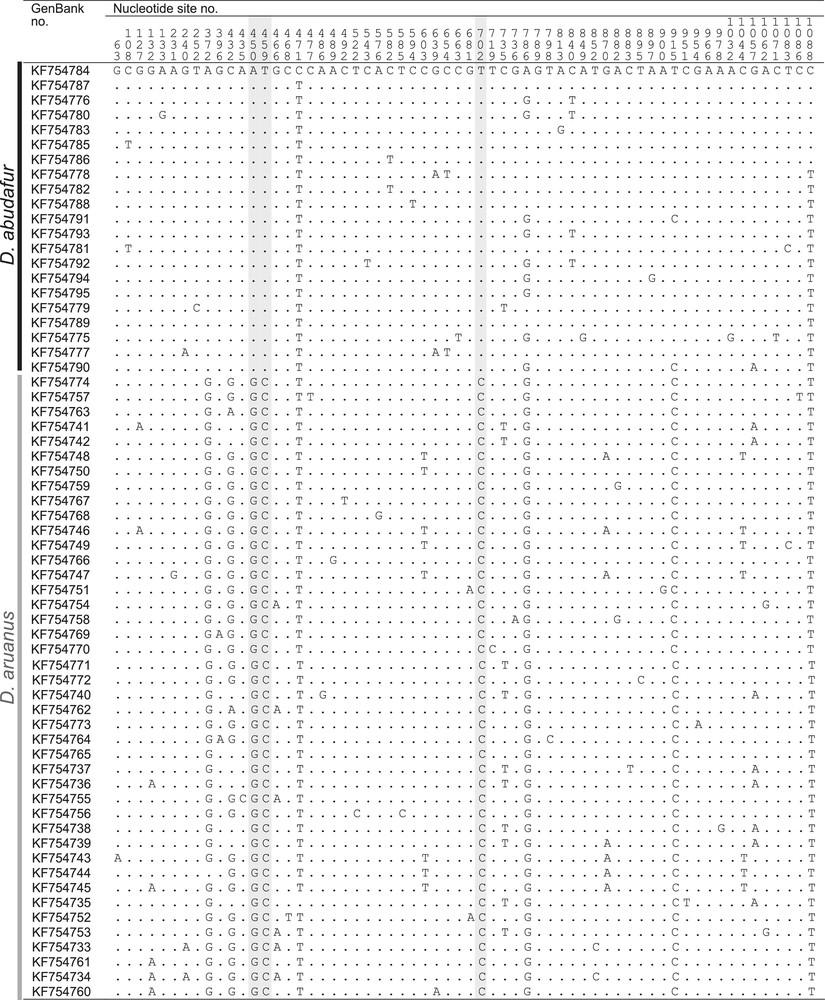
Variable nucleotide sites in cytochrome b gene sequences of humbug damselfish, Dascyllus abudafur and D. aruanus. Nucleotide sites numbered from the homologous first nucleotide of the gene in Salmo salar (GenBank EU492281) and mutations marked using S.-Y.V. Liu et al.’s [26] sequence of the most common haplotype from the Red Sea, RS02 (GenBank KF754784), as reference. Haplotypes ordered by location of first occurrence, from West to East (Supplementary material, Table S3). Highlighted grey: nucleotide site diagnostic between D. abudafur and D. aruanus; dot: nucleotide identical to reference (Appendix A).
Two main nuclear forms, Indian and Pacific, were also determined from microsatellite genotypes [26]. The distribution of these two forms is presented Fig. 1B. Genetic differences at nuclear loci were highly correlated with those at the mitochondrial locus (Appendix A).
The average score of pigmentation intensity was 2.51 ± 0.68 in the Indian Ocean sample (n = 49) and 0.08 ± 0.27 in the Pacific Ocean sample (n = 103). It was 2.61 ± 0.63 in the Indian mitochondrial form (n = 28) and 0.20 ± 0.40 in the Pacific mitochondrial form (n = 46). The null hypothesis of no differences in pigmentation patterns between the two oceans was rejected (χ2 test; 3 df; P < 10−28), and so was it between the two mitochondrial forms (χ2 test; 3 df; P < 10−13). A majority (62%) of individuals from the Indian Ocean exhibited a blackish blotch at the basis of the caudal fin and no one was unpigmented. Conversely, a majority (92%) of Pacific Ocean individuals had an unpigmented caudal fin, the remainder (8%) being slightly pigmented (details in Supplementary material, Table S2). All individuals of the Indian mitotype and for which a photograph was available, except two, possessed a blackish or solid-grey blotch at the basis of the caudal fin. For the two exceptions, from Europa Island, the basis of the caudal fin was light-grey. Conversely, the caudal fin of individuals of the Pacific mitotype was entirely white in most cases, or with a light-grey basis. Individuals sampled in the presumed zone of transition, i.e., Lombok Strait (n = 23) exhibited a variety of pigmentation patterns, from white to blackish (Supplementary material, Table S2). A specimen exhibiting the typical pigmentation pattern of the Indian Ocean form is presented Fig. 2A, while a typical specimen of the Pacific form is presented Fig. 2B.
4 Discussion
Under K. de Queiroz's unified species concept [31], species are defined simply as separately evolving metapopulation lineages. K. de Queiroz [31] and other authors [32,33] have emphasized the fundamental distinction between the definition of species, and species delimitation. The latter concerns properties of species, which include, e.g. morphological distinction, niche separation, reproductive isolation, and monophyly. These properties, one by one or altogether, constitute the lines of evidence researchers in practice may have access to, to hypothesize the existence of a species [31–33].
Several of the expected properties of species characterize the Indian Ocean versus the Pacific Ocean forms of humbug damselfish. The mitochondria of humbug damselfish from the Indian Ocean belong to a single lineage sister to that from the Pacific Ocean: the two forms are reciprocally monophyletic [25–27]. No location with mitotype (Indian vs. Pacific) polymorphism was observed, except at the western shore of Lombok Strait. The transition zone between the two mitochondrial forms lies east of the Central Java Sea and west of Lombok. A phylogeographic partition into two forms was also evident from allelic frequencies at nuclear loci, similar to that at the mitochondrial locus [26]. The location of the nuclear transition zone, which could not be determined with as much precision as the mitochondrial one, lied east of Madagascar and west of the central part of the Coral Triangle. The fact that population genetic differences at nuclear loci were highly correlated with differences at the mitochondrial locus indicates a common history of divergence, between the two forms, for the nuclear and mitochondrial genomes. The distribution of these two genetically separate forms of the humbug damselfish thus appears to be either parapatric, or slightly overlapping, in a narrow transition zone located at the western edge of the Coral Triangle. The two forms were also characterized by distinct pigmentation patterns, a criterion of considerable importance to taxonomists working on reef fishes (e.g., [6,9,13]). The two forms thus fulfil the criteria of morphological and genetic distinctness, which in turn allows us to consider them as distinct evolutionary units, hence separate species under the unified species concept [31]. In addition, their apparently parapatric distribution suggests some degree of reproductive isolation [34], which is another property of separate species [31].
In C. Niebuhr's posthumous publication of the notes taken by P. Forsskål during his naturalist expedition to the Red Sea, a fish from Jeddah locally named abu dafur was mentioned in the following terms: “Chaetodon abu dafur Arab. est charactere Aruani Linn. Albus, fasciis tribus nigris: spinis Dors. 12. A. 2. Djiddæ habitat, rarior, inter corallia. Spith. Longit.” [35]. It is possible that P. Forsskål considered this fish to be identical with C. Linnaeus’ Chaetodon aruanus [36]. D. abudafur (Forsskål) was subsequently synonymized with D. aruanus (Linnaeus) [37,38]. Epithet aruanus has since then been maintained as the single valid one to designate both the Indian and the Pacific forms of the humbug damselfish [29,39–42]. C. Linnaeus writes that D. aruanus “Habitat in Indiis” [43]. W.N. Eschmeyer precisely indicates the type locality to be Aru Islands, Moluccas [42]. Since the Aru Islands are located within the distribution range of the Pacific mitotype, epithet aruanus is here kept to designate the Pacific form of the humbug damselfish. No description of the Indian Ocean form of humbug damselfish is known before P. Forsskål's [42]. Hence, the Indian Ocean form should be renamed D. abudafur.
There are numerous examples of allopatric lineage diversification among marine populations on either side of the Coral Triangle ([1,2] and references therein). However, ecological factors that have led to lineage diversification, and those that maintain it, are difficult to identify because of the usual difficulty to distinguish contemporary, extrinsic barriers to larval connectivity, from internal barriers to gene flow, i.e. reproductive isolation [44,45]. Transition zones are privileged situations for addressing such questions, for they offer the possibility to explore the apportion of the various mechanisms, passive vs. active, involved in speciation in marine organisms [46–48]. Investigating the population genetic structure of humbug damselfish populations across the transition zone between D. abudafur and D. aruanus is thus warranted. Phylogeographic surveys similar to those undertaken in D. aruanus ([25,26]; present study) are needed to assess the taxonomic status of separate genetic forms within a nominal species, in all other cases where a geographic break has been identified.
5 Taxonomy
Dascyllus abudafur (Forsskål 1775), resurrected species.
5.1 Previous denominations
Chaetodon abu dafur[35]; Pomacentrus aruanus[37]; D. aruanus ([38,39,49,50]; other references in [42]).
5.2 Diagnosis
A Dascyllus of white pigmentation with three transversal black bands [35]. Caudal fin bilobate. Possesses a grey to blackish blotch at the basis of the caudal fin that distinguishes it from its Pacific Ocean sister-species, D. aruanus [43], whose tail is generally entirely white. Also distinct from D. aruanus by its cytochrome b gene sequence: nucleotide sites nos. 450, 459 and 702 of the gene are diagnostic between the two species (Fig. 3). At these three sites, D. abudafur has the triplet (A, T, T) whereas D. aruanus has (G, C, C).
5.3 Distribution
Indian Ocean, from the Red Sea to the Sunda Shelf.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgements
We are grateful to G. Bernardi, J.-D. Durand, Husni, G. Mou-Tham and S. Planes for participating in the collection of samples; J. Bertrand for help with sequencing analysis; and an anonymous reviewer for helpful suggestions. Access to 18th- and 19th-century books was through the Biodiversity Heritage Library website (http://www.biodiversitylibrary.org). Sampling in the Mozambique Channel was done under Program CNRS-INEE/TAAF “Îles Éparses” (PhylIP project: PB). Sampling in Dongsha was sponsored by the Oceanography Institute of NTU (WJC). Sampling in West Papua was done during LIPI/Ekspedisi Widya Nusantara on board RV Baruna Jaya VIII in November–December 2007 (PB). Sampling at Madang was part of the Niugini 2012 biodiversity expedition (WJC). Sampling in New Caledonia was during the Resicod workshop sponsored by the Fondation de recherche pour la biodiversité (M. Kulbicki, R. Myers, P. Pruvost and PB). WJC acknowledges grant support from Ministry of Science and Technology in Taiwan (MOST 101-2611-M-002 -016 -MY3). Also funded in part by IRD UR 227 and IBRC. PB designed the study. PB, AS, CF, WJC contributed reagents and/or materials and/or analysis tools. PB, WJC analysed and interpreted the data. PB wrote the paper.
Appendix A
Correlation of pairwise θ estimates at the mitochondrial locus against homologous θ estimates at 7 microsatellite loci in humbug damselfish (from Supplementary Tables S2 and S3 of [26]). Mantel's test of the correlation between the two matrices: Z = 1.770; r = 0.869; P < 0.001.


