1 Introduction
Kelp (Haidai in Chinese) is an economically important brown macroalga, which was introduced in China from Japan in 1927 and widely cultured in Shandong, Liaoning and Fujian Provinces [1]. It was widely used in medicine, in the food and chemical industries among others, occupying an important place in the mariculture industry of China. Up to 2013, the culturing area of S. japonica in China reached 37,282 hectares, with a total production of 1,017,737 tons [2]. China has ranked the first in the world for many years in the culture area and annual yield of kelp.
The life history of kelp consists of a diploid sporophyte generation and a haploid gametophyte generation, alternating with each other. Sporophyte generation could not be conserved as germplasm as its large frond and can not be long-term preserved indoor. Fang et al. [3] found that the male and female gametophytes of kelp can be isolated and propagated in vitro independently under continuous lighting (gametophyte cloning). In the past, gametophytes can only be acquired from mature sporophytes, a process that was limited by the season. The establishment of the kelp gametophyte clone culture technique could provide the gametophytes as laboratory research materials at any time throughout the year [4]. Since then, cloning kelp gametophytes has evolved as an effective way for long-term preservation of the kelp germplasm. The technique has facilitated the preservation of individual genetic strains of Saccharina and promoted the subsequent breeding of elite varieties.
Cloning kelp gametophytes has promoted kelp hybridization breeding. The innovation of Saccharina gametophyte cloning and hybridizing methods has brought Saccharina breeders the opportunity of breeding elite Saccharina varieties and Saccharina hybrids by crossing gametophyte clones [5,6]. As the earliest country of kelp genetic improvement, more than 20 elite varieties have been bred and applied to production since the late 1950s [7]. In recent years, a set of kelp hybrids and hybrid derived varieties have been bred and applied. The outstanding representatives of kelp hybrids and hybrid derived varieties include, Dongfang No. 2 [8], Dongfang No. 3 [9], Dongfang No. 6 [7], 901 [10,11] and Dongfang No. 7 [12].
At present, cloned gametophytes are the entities of germplasm conserved indoors and the parental resources of the Saccharina variety and hybrid breeding. A few germplasm stocks of Saccharina gametophyte clones have been constructed. A rich collection of Saccharina gametophyte clones were collected and preserved indoors. Various Saccharina gametophytes with specific phenotypic characteristics were obtained and applied to breeding. However, little is known about the genetic background of the Saccharina gametophytes at the DNA levels. In addition, the performance of hybrids is associated with the differentia between parental gametophytes [13]. Except for those from sporophytes with desirable traits, we have to determine the genetic difference among gametophyte clones so that the differentia can be identified and utilized for parental gametophytes selection of hybrid kelp breeding. In order to effectively manage and exploit Saccharina gametophyte clones, the genetic diversity and the phylogenetic relationships among 48 Saccharina germplasms were assessed.
Diverse molecular markers have been used widely to evaluate the genetic variation of a wide range of species, which included, for example, simple sequence repeat (SSR), RAPD, ISSR, and amplified fragment length polymorphism – AFLP [14–18]. For Saccharina, 18 microsatellites have been developed and used to determine the genetic diversity of gametophyte clones originated from Laminaria japonica and L. longissima [19,20]; 18 other microsatellites have also been used to determine the genetic distance between parental gametophyte clones of 14 Laminaria hybrids and predicting the heterosis of Laminaria hybrids [13]. In addition, AFLP, RAPD and ISSR have been adopted in analyzing the genetic distance between gametophyte clones [21], fingerprinting gametophytes [22], and assessing the genetic diversity [23]. These markers are PCR-based and widely used in plant species for identification, phylogenetic analyses, population studies, and genetic linkage mapping.
Of the molecular markers mentioned above, ISSR, which was derived from SSR, amplified the specific region between two microsatellite motifs, and it does not require prior knowledge of the DNA sequence for primer design [23,24,25]. Compared with SSR, ISSR proved to be abundant, highly polymorphic, informative, and efficient, even in closely related genotypes among selected samples in the pre-experimental part of this study. The AFLP technique involves the high-throughput detection marker system of electrophoresis, but enzyme digestion and the detection system of AFLP molecular markers are complex to operate, and high requirements on quality of genome DNA are needed. The RAPD technique has been successfully employed in genetic diversity studies of some species due to the simplicity, low cost and non-requirement of DNA sequence information prior to application [24,26]. Both RAPD and ISSR markers have proved to be reliable, easy to generate, inexpensive and versatile set of markers that rely on repeatable amplification of DNA sequence using single primer.
Overall consideration, ISSR and RAPD were adopted for evaluating the genetic relationship of a selected collection of Saccharina in the present study. The aim of the study was to assess the genetic background in order to understand the relationship of the selected varieties (lines), and to identify genetic differentia of selected gametophytes and to screen the candidate parental gametophytes that were used for hybrid kelp breeding test. The two marker systems (ISSR and RAPD) have also been compared for their applicability to the selected Saccharina gametophytes population. In addition, the results also indicated that the two marker systems can be used for kelp core germplasm selection.
2 Materials and methods
A total of 48 gametophytes either isolated from 17 varieties (or lines) of Saccharina (maintained by independent sporophyte seedling raising) or collected worldwide (Table 1, Fig. 1) were analyzed. These gametophytes preserved in The Germplasm Repository of National Engineering Science Research & Development Center of Algae and Sea Cucumbers of China. At least one female and one male gametophyte were selected to represent a variety or a line except for the two wild lines (6, 9), Rongyuanyuan-1 (3, male) and Yuanza (11, male).
List of the kelp gametophytes used in this study.
| Gametophyte | Source | Sporophytic characteristics |
| 1,1 | 901 with high algin content | Triangle base, circular stipe, light brown blade with obvious vertical channel, with more algin content than 901 |
| 21, 22, 21, 22 | 901 | Triangle base, circular stipe, long and thin blade with obvious vertical channel, high growth rate and resistant to high temperature |
| 3 | Rongyuanyuan-1 | Flourishing holdfast, circular base, dark brown blade and high water content |
| 41, 42, 41, 42, 43 | Zaohoucheng No.1 | Flourishing holdfast, little flat stipe, circular base, dark brown and thick blade and early in sporangium development |
| 5, 5 | Haiza | Broad and thin blade |
| 61, 62, 63 | Wild strain from Changdao | Flourishing holdfast, long and narrow and dark brown blade |
| 7, 7 | Feijicai | Flourishing holdfast, circular base, flat and thick stipe |
| 81, 82, 81, 82 | Penglaicai | Flourishing holdfast, partial circular stipe, triangle base, wide middle part and narrow edge |
| 9 | Wild strain from Rishiri Island | No description |
| 101, 102, 101, 102 | Hanguo | Flourishing holdfast, circular base, flat stipe, highly tolerant to high seawater temperature and solar irradiation, flexible in blade texture, late in sporangium development and low in blade water content |
| 11 | Yuanza No. 10 | Flourishing holdfast, flat stipe, triangle base, brown blade with fossa |
| 121, 122, 12 | Benniu | Flourishing holdfast, flat stipe, wide in blade with obvious vertical channel and few spots |
| 131, 132, 13 | Haifeng | Flourishing holdfast, narrow and flat stipe, triangle base, light brown blade with obvious channel |
| 141, 142, 141, 142 | Lianza No. 1 | Flat stipe, circular base, brown blade, wide in blade shape, tolerant to high seawater temperature and late in sporangium development |
| 151, 152, 15 | Rongfu | Flourishing holdfast, flat stipe, triangle base, light brown blade with obvious vertical channel and few spots, tolerant to high seawater temperature |
| 16, 16 | Haike | Flourishing holdfast, triangle base, wide blade, no obvious middle part |
| 171, 172, 171, 172 | Wild strain from Tuoji Island | Flourishing holdfast, triangle base, brown wide blade with obvious vertical channel |

Geographical location and sampling sites. The white circles show the detailed sites.
Genomic DNA was extracted from 0.1 g (dry weight) of gametophytes using the plant genomic DNA extraction kit (Tiangen, China). DNA quality and quantity were checked through 1.0% agarose gel electrophoresis and spectrophotometry, respectively. DNA was diluted to 40 ng/μL as templates and stored at –20 °C.
Fifty ISSR random primers were synthesized by Sangon (Shanghai, China) and pre-screened in a part of gametophytes, with primers that produced reproducible, clear and polymorphic bands were chosen to amplify all gametophytes. PCR was carried out in a Bio-Rad Thermal cycler T-100 and a 20-μL reaction volume containing 40 ng of template DNA, 1 × polymerase buffer (Promega, USA), 1.2 mmol/L of Mg2+, 0.2 mmol/L of dNTP (each), 1 μmol/L of ISSR primer and 0.25 U of Taq DNA polymerase (Promega, USA). By pre-denaturation at 95 °C for 3 min, followed by 38 cycles of denaturation at 95 °C for 45 s, annealing at 52 °C for 45 s, and extending at 72 °C for 2 min, and a final extension at 72 °C for 10 min. Fifty RAPD random primers were synthesized by Sangon (Shanghai, China) and were pre-screened. The primers that produced reproducible, clear and polymorphic bands were chosen. PCR was carried out in a Bio-Rad Thermal cycler T-100 and a 20-μL reaction volume containing 40 ng of template DNA, 1 × polymerase buffer (Promega, USA), 1.0 mmol/L of Mg2+, 0.2 mmol/L of dNTP (each), 2 μmol/L of ISSR primer, and 0.25 U of Taq DNA polymerase (Promega, USA). By pre-denaturation at 95 °C for 5 min, followed by 40 cycles of denaturation at 95 °C for 30 s, annealing at 37 °C for 1 min, and extending at 72 °C for 2 min, and a final extension at 72 °C for 10 min. The amplified product was separated on 1.5% agarose gel, and recorded with a Tanon 2500 gel documentation system.
ISSR and RAPD bands were recorded as presence (1) or absence (0) in a gametophyte. The percentage of polymorphism was calculated by dividing the polymorphic bands by the total and then multiplied with 100. The polymorphism information content (PIC) of each ISSR and RAPD primer was calculated using the formula: PIC = 1–∑(Pij)2, where Pij is the frequency of the ith pattern revealed by the jth primer summed across all patterns revealed by the primers [27]. Similarity matrices were constructed according to simple matching similarity coefficients using NTSYS-pc (numerical taxonomy and multivariate analysis system) software package [28]. Genetic similarity was calculated from the simple matching similarity coefficients for all the 48 gametophytes of Saccharina, considering ISSR and RAPD approaches individually as well as together. Cluster analysis was performed on the basis of a genetic similarity matrix and the resulting similarity coefficients were used for constructing a dendrogram using UPGMA with the SAHN module of NTSYS-pc. The Mantel matrix correspondence test was used to test the significance of the correlation between matrices obtained from two marker systems [29]. Eigen values and eigen vectors from a similarity matrix were used for calculating PCA values using NTSYS-pc software.
3 Results
3.1 Genetic diversity revealed by ISSR and RAPD markers
A total of 17 ISSR primers produced 262 bands, of them, 256 were polymorphic, accounting for a polymorphism of 97.12%. The number of amplified bands varied between 7 (primer 844) and 22 (primer 823, 834, 857). The average number of polymorphic bands per primer was 15.06. PIC value ranged from 0.781 (primer 844) to 0.933 (primer 857), with an average of 0.879 (Table 2). In case of RAPD analysis, of 50 RAPD random primers, 16 yielded a clear and reproducible band pattern. A total of 355 RAPD bands were produced, of them 352 were polymorphic, accounting for 98.61%. The number of amplified bands varied between 11 (primer S2114) and 38 (primer S51) with an average of 22. PIC value ranged from 0.780 (primer S112) to 0.967 (primer S51) with an average of 0.909 (Table 3). The represented agarose gel electrophoresis pictures of PCR fragments were shown (Fig. 2).
Data for ISSR primers used for analyzing 48 gametophytes of kelp.
| Primer | Sequence | Size Range |
No. of bands | No. of polymorphic bands | Percentage | PIC |
| 808 | (AG)8C | 340–967 | 12 | 12 | 100 | 0.852 |
| 810 | (GA)8T | 210–1850 | 11 | 10 | 90.91 | 0.783 |
| 811 | (GA)8C | 210–1400 | 17 | 17 | 100 | 0.895 |
| 815 | (CT)8G | 450–2923 | 13 | 13 | 100 | 0.849 |
| 823 | (TC)8C | 250–2000 | 22 | 21 | 95.45 | 0.899 |
| 834 | (AG)8YT | 250–2800 | 22 | 22 | 100 | 0.932 |
| 835 | (AG)8YC | 180–800 | 13 | 12 | 92.31 | 0.889 |
| 844 | (GA)8YT | 470–2400 | 7 | 6 | 85.71 | 0.781 |
| 848 | (CA)8RG | 160–2000 | 20 | 19 | 95.0 | 0.909 |
| 855 | (AC)8YT | 180–1400 | 15 | 15 | 100 | 0.920 |
| 857 | (AC)8YG | 245–1600 | 22 | 22 | 100 | 0.933 |
| 859 | (TG)8RC | 250–2000 | 19 | 19 | 100 | 0.917 |
| 864 | (ATG)6 | 240–1800 | 16 | 16 | 100 | 0.899 |
| 880 | GGA(GAG)2AGGAG | 180–1270 | 12 | 11 | 91.67 | 0.893 |
| 884 | HBHA(GA)6G | 400–1500 | 9 | 9 | 100 | 0.865 |
| 889 | BHBG(AG)6A | 200–1100 | 18 | 18 | 100 | 0.908 |
| 890 | (GGAGA)3 | 362–1285 | 14 | 14 | 100 | 0.831 |
| Total | 262 | 256 | ||||
| Average | 15.41 | 15.06 | 97.12 | 0.879 |
Data for RAPD primers used for analyzing 48 gametophytes of kelp.
| Primer | Sequence | Size range | No. of bands | No. of polymorphic bands | Percentage | PIC |
| S30 | GTGATCGCAG | 350–3000 | 21 | 21 | 100 | 0.886 |
| S31 | CAATCGCCGT | 200–2700 | 26 | 26 | 100 | 0.937 |
| S51 | AGCGCCATTG | 280–2400 | 38 | 38 | 100 | 0.967 |
| S55 | CATCCGTGCT | 350–2200 | 19 | 19 | 100 | 0.928 |
| S60 | ACCCGGTCAC | 200–2900 | 26 | 26 | 100 | 0.941 |
| S80 | ACTTCGCCAC | 300–2100 | 21 | 21 | 100 | 0.907 |
| S104 | GGAAGTCGCC | 280–2500 | 36 | 36 | 100 | 0.964 |
| S112 | ACGCGCATGT | 300–1800 | 12 | 11 | 91.67 | 0.780 |
| S1501 | CTACGGCTTC | 450–3300 | 21 | 20 | 95.24 | 0.881 |
| S1513 | GGCTTGGCGA | 250–2400 | 22 | 22 | 100 | 0.939 |
| S2110 | GTGACCAGAG | 200–3750 | 21 | 21 | 100 | 0.910 |
| S2111 | GACGACCGCA | 350–1650 | 22 | 22 | 100 | 0.923 |
| S2114 | CCGCGTTGAG | 350–2000 | 11 | 10 | 90.91 | 0.818 |
| S2115 | ACGCGAACCT | 170–2500 | 26 | 26 | 100 | 0.946 |
| S2116 | AGGGTCCGTG | 370–2500 | 17 | 17 | 100 | 0.920 |
| S2118 | AGCCAAGGAC | 300–2700 | 16 | 16 | 100 | 0.896 |
| Total | 355 | 352 | ||||
| Average | 22.19 | 22 | 98.61 | 0.909 |

An ISSR PCR product amplified by primer 811. M, DNA marker. Lanes 1 to 15 represent partial samples; b RAPD PCR product amplified by primer S55. M, DNA marker. Lanes 1 to 15 represent partial samples.
3.2 Dendrogram analysis by ISSR and RAPD markers
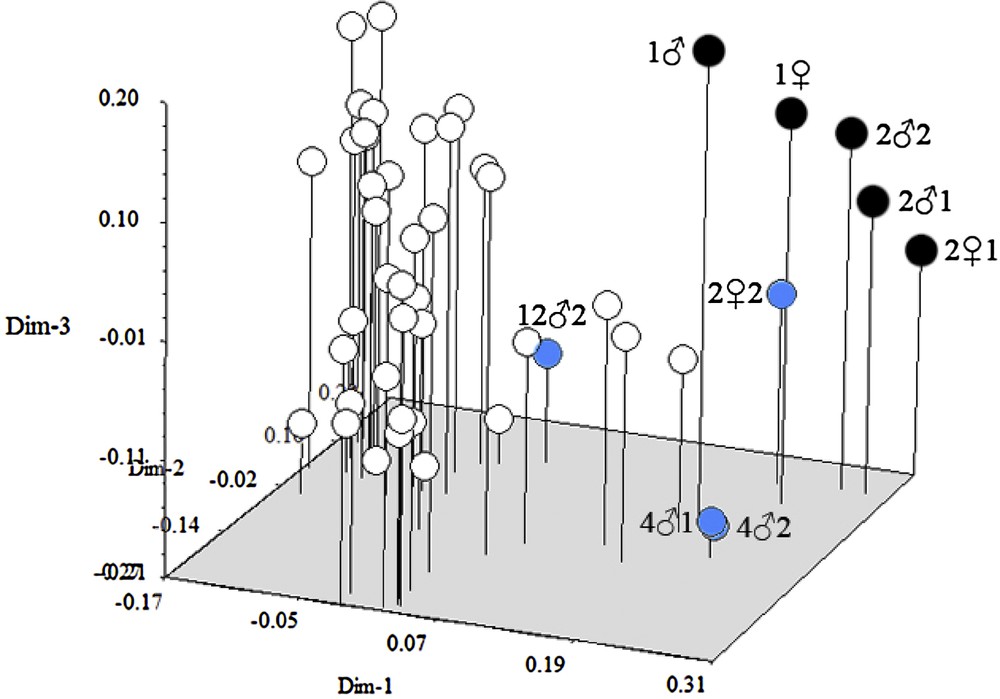
Based on ISSR markers, the similarity coefficients ranged from 0.568 to 0.885. These values were used to construct a dendrogram using UPGMA. In the ISSR-based dendrogram, the 48 gametophytes clustered three main groups at a cutoff value of 0.68 (Fig. 3). A relationship among the 48 gametophytes individuals was also detected by PCA based on ISSR data. Gametophytes grouped within the same cluster in the dendrogram occupied the same group positions in the three-dimensional scaling (Fig. 6). Group I included 1, 1, 21, 22 and 21 (Fig. 3). The five individuals were also clustered together in the three-dimensional scaling; they were labeled and shown in black circles. Group III included 22, 41, 42, which were also clustered together and were labeled and shown in blue circles. The remaining 40 gametophytes clustered in Group II were also gathered together in the three-dimensional scaling and shown in white circles (Fig. 6). Based on RAPD markers alone, the similarity coefficients ranged from 0.670 to 0.873. In the RAPD-based dendrogram, the gametophytes formed three clusters at a cutoff at 0.74 (Fig. 4). The three dimensional scaling of 48 gametophytes based on RAPD data was shown (Fig. 7). The gametophytes clustered in Group I were labeled in white circles. Group II was labeled in black circles. The extra gametophyte, 122, was shown in blue circle (Fig. 7).
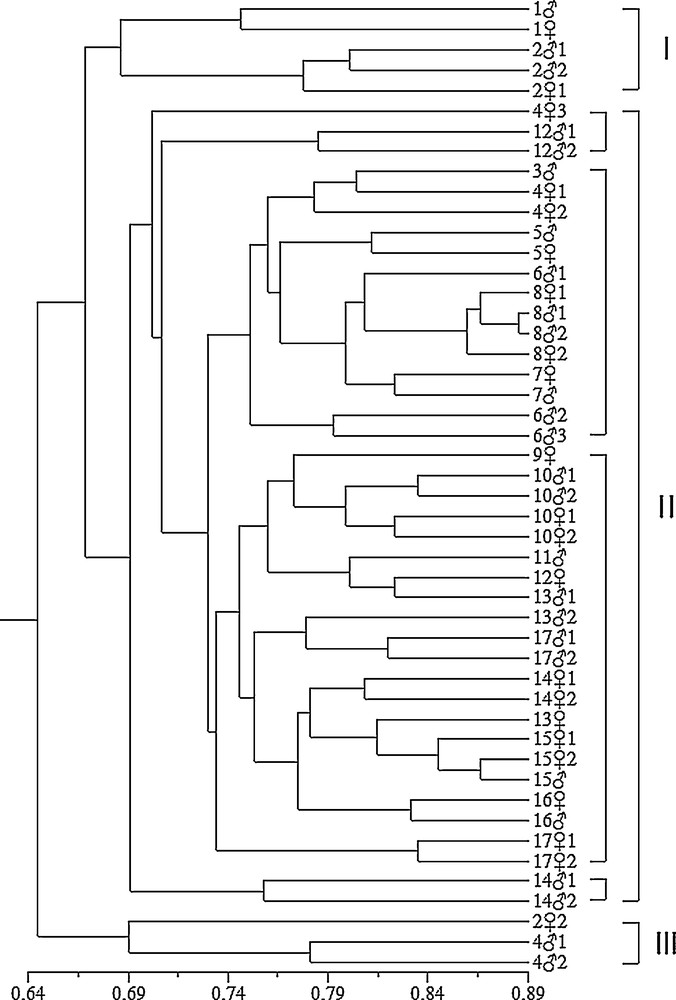
UPGMA cluster analysis of 48 gametophytes with similarity coefficient of ISSR.

Three-dimensional plots of 48 gametophytes by PCA based on ISSR data.
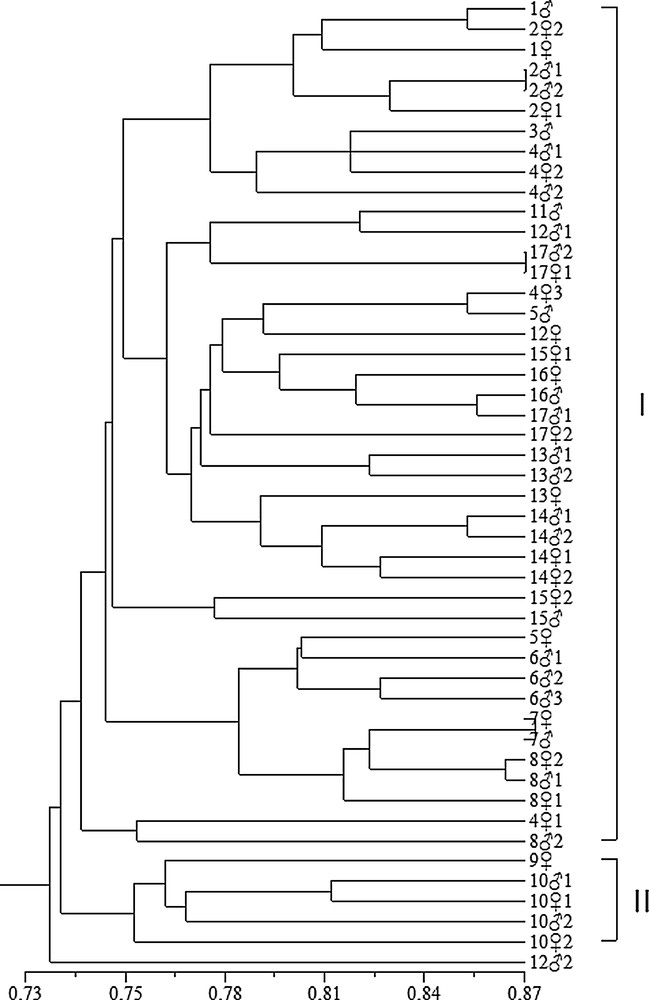
UPGMA cluster analysis of 48 gametophytes with the similarity coefficient of RAPD.

Three-dimensional plots of 48 gametophytes by PCA based on RAPD data.
Based on both the marker systems, the similarity coefficients ranged from 0.667 to 0.862. Cluster analysis performed from combining data of both markers generated a dendrogram separating the gametophytes into three main clusters at a cutoff value at 0.72 (Fig. 5). Relationship among all the 48 gametophytes was also shown by PCA based on pooled RAPD and ISSR data (Fig. 8). The individuals in Group I were labeled in black circles. Group II was labeled in white circles. Group III was shown in blue circles (Fig. 8).

Dendrogram of 48 gametophytes based on the analysis of pooled ISSR and RAPD.
Three-dimensional plots of 48 gametophytes by PCA based on ISSR and RAPD data.
Our PCA results revealed that the gametophytes belonging to a particular cluster were grouped together in a PCA plot. The three-dimensional plot positions were basically consistent with the clustering pattern of the dendrogram (Figs. 6–8), which indicated substantial genetic diversity among gametophytes. It is evident from data of both marker systems that gametophytes are fairly dispersed on PCA plots, which reflects a good genetic base.
3.3 Comparison between two markers
The cophenetic correlation coefficient was positively correlated to the Mantel test statistics in both ISSR and RAPD in the present study. The correlation coefficient corresponded to a good fit in ISSR (matrix correlation: r = 0.78), while a moderate fit was observed in RAPD (r = 0.64). When the similarity matrices generated using ISSR and RAPD markers were compared, a value of r = 0.62, P > 0.001, indicated a moderate correlation between data generated by both marker systems.
3.4 Molecular-assisted selection of parental gametophytes for hybrid breeding
The hybrid kelp obtained from male and female gametophytes of different sporophytes, with great differences not only in phenotype but also in genotype, could show improved trait and high yield. Genetic relationship analysis could assist in parental gametophytes selection in hybrid breeding. In this study, the sporophytes of Peilaicai (8) and Haike (16) showed large difference of phenotypic properties (Table 1). Genetic analysis indicated that the gametophytes from the Peilaicai and Haike showed genetic stability within the varieties respectively, while the distant relationships were observed between gametophytes from Peilaicai and Haike (Fig. 3). The similar combinations of 901 with high algin content (1) and Lianza No.1 (14), Hanguo (10) and Lianza No.1 (14) also showed different shape characteristic and genetic distant relationships. These results provided assistance for parental gametophytes selection for hybrid breeding; the combinations could be candidate parental gametophytes for the hybrid breeding test.
4 Discussion
Identification of genetic relationships or genetic diversity is helpful to understand the genetic background of the germplasm resources and provide genetic guidance for parental gametophytes selection for hybrids kelp breeding. In this study, two marker systems, ISSR and RAPD, were used to evaluate the genetic diversity and the genetic relationships among 48 gametophytes from 17 varieties (or lines). Comparison of genetic similarity coefficients of both ISSR and RAPD markers showed that the former ranged from 0.57 to 0.89, while the latter varied from 0.67 to 0.87. Thus, both marker systems showed polymorphisms and large variability, and could distinguish the individuals clearly to a certain extent. The results indicated that high diversity among 17 varieties (or lines) and assisted in parental gametophytes selection for hybrid kelp breeding.
The selected ISSR primers (Table 2) in this study indirectly indicated the existence of microsatellite regions of (TC)n, (AC)n, (CA)n, which were found in the S. japonica genome [19]. Most of them were also found in Saccharina gametophytes [23,25]. ISSR are highly reproducible due to their primer length and very stringent annealing temperature [30,31]. RAPDs are easy, fast and sensitive [22]. These two quick and simple marker systems were widely used for the studies of phylogeny and genetic diversity of Saccharina. An average of 15.41 loci per ISSR primer (Table 2) and 22.19 loci per RAPD primer (Table 3) were obtained in this study, which were considerably > 12.3 loci per ISSR primer in genetic variation analysis of 100 selected Saccharina gametophytes [25] and 13.0 loci per RAPD primer in germplasm characterization of 33 differently selected Saccharina gametophytes [22]. As for Saccharina, many studies have been carried out with ISSR and RAPD to identify the cultivars of Saccharina [22,23,25,32]. ISSR markers were utilized to investigate the genetic variation between male and female gametophyte populations of Saccharina [25]. He et al. [33] have assessed the germplasm of Laminaria with the RAPD method. Isoenzyme and RAPD technique were applied to analyze the genetic diversity of gametophytes of different cultivars in China [34]. The former studies indicated that the RAPD and ISSR markers are stable and could be used to evaluate the germplasm resources. In this study, the loci of ISSR and RAPD shared by most gametophytes have proved to be reproducible and stable through repeated tests. The results indicated that both ISSR and RAPD markers were effective to assess the selected gametophytes, while matrix correlation of ISSR marker system (r = 0.78) was a little better than that of RAPD marker system (r = 0.64). In addition, both DNA marker methods identified specific amplified bands for varieties (or lines), and a higher number of variety-specific loci was revealed by RAPD markers than by ISSR markers. These specific alleles would be converted to co-dominate sequence-characterized amplified region (SCAR) markers in further studies.
The dendrogram clusters could show the genetic relationships between varieties or lines. Most of the varieties or lines could be distinguished from each other and showed high genetic variation (Fig. 3, Table 3). Our results were consistent with those of the previous study using isozymes [34], RAPD [22,33,34] and ISSR [23,25]. The common cultivars in China, Haiza (5), Peilaicai (8), Rongfu (15) and Haike (16) showed a low level of genetic variation within variety (or line) (Fig. 3), which indicated their genetic stability, while the other common cultivars including 901 (2), Zaohoucheng No. 1 (4), Benniu (12), Haifeng (13), Lianza No. 1 (14) showed high variations within variety (or line); for example, the male and female gametophytes of Zaohoucheng No. 1 (4) clustered into different groups (Fig. 3). The genetic variations within a variety were probably attributable to the possible following reasons: firstly, there may exist some genetic variation in order to adapt to the nature environment, and the subsequent intense artificial hybridized selections might increase the genetic diversity within a variety. Secondly, there were many varieties (lines) cultivated at the same time, there occasionally mixed with each other if their phenotype were not very evident. Finally, there may be some mutations occurring in the process of long-term preservation after having been isolated from sporophytes, which remains to be confirmed [23,25]. On the other hand, the germplasm of high-diversity varieties could be potentially used to broaden the genetic base of kelp cultivars, and could be explored for novel genes that can be beneficial to the improvement of economically important traits.
Involving the sampling sites, the four geographical locations, Fujian and Shandong provinces of China, Japan and Korea, were included. Wild strain from Rishiri Island (9) and four gametophytes (101, 102, 101, 102) of Hanguo line have close relativeness degrees in both ISSR and RAPD dendrograms (Figs. 3 and 4). 9 was collected from wild kelp strain grown on the coast around Rishiri Island in Japan in 2001 and preserved till now. The four gametophytes of Hanguo kelp were collected from cultivated strain which originated from the coast of Korea in 2007. Lianza No. 1 (14) was collected from Fujian province, located in the southeast of China. Lianza have close genetic relationship with Haifeng (13) and Rongfu (15), which were collected from Shandong province, and they all clustered in Group II together with other cultivar varieties (lines) (Fig. 3). The other two wild strains, the one from Changdao (6) and that from Tuoji Island (17), were also included in genetic diversity analysis. There was no obvious geographical variation in this study.
The parents used for hybrid kelp breeding were gametophyte clones that do not have properties comparable with those of sporophytes. The selection of parent gametophytes was only based on the phenotype of the originated sporophytes. The molecular marker analysis of the genetic relationship of the gametophytes facilitates the selection on the basis of the molecular levels. Heterosis could be obtained from the large differentia of the phenotype and far relation of genotype [13]. In this study, several combinations (such as Peilaicai and Haike, 901 with high algin content and Lianza No. 1, Hanguo and Lianza No. 1) were recommended as the candidate parents for hybrid kelp breeding. Molecular-assisted selection could avoid field evaluation of a large number of gametophytes combinations and accelerate the breeding process [8].
The varieties or lines cultivated in China mainly belong to Saccharina japonica bred through multiple generation of hybrid or selfing selection. The genetic background is relatively complex. The kelp gametophytes were preserved in liquid phase with low temperature. There is occasionally miscellaneous bacteria contamination or bad growth status of some samples. Therefore, in the germplasm collection process, the gametophytes were isolated from several different mature sporophytes and cultivated respectively for each variety or line. This led to a maybe repeated preserved germplasm, and, along with long-term preservation, generated more cost of material and financial resources. So, the screening of core collection of kelp should be carried out on the level of variety or line. The results of genetic relationship analysis indicated that RAPD and ISSR markers could be used for core germplasm screening for kelp gametophytes of different varieties.
Determination of genetic diversity and genetic relationship is a primary step for assessing and efficiently utilizing the available Saccharina germplasm resources [17,32]. The results in this study help us to understand the relationship among the 48 gametophytes from 17 varieties (or lines). It also provided guidance for parental gametophytes selection for kelp hybrid breeding on the level of molecular level, which indicated that molecular-assisted breeding is feasible. The RAPD and ISSR markers could be used to carry out core germplasm collection as they could detect the variation within variety or line. The specific amplified bands were found by RAPD and ISSR marker respectively, they would be used to develop the SCAR marker for molecular identity of different varieties or lines in a further study.
Disclosure of interest
The authors declare that they have no competing interest.
Acknowledgments
This study was funded by National Supporting Program of Science and Technology of China (2012BAD55G01), the National High Technology Research and Development Program (863 Program) of China (2012AA10A406), and the Municipal Science and Technology Research and Development Project of Yantai (2013LGS002), and the earmarked fund for Modern Agro-industry Technology Research System in Shandong Province (SDAIT-26-11).
We thank Professor Guanpin Yang for his help on manuscript writing and language change. We also thank editors and reviewers for their patience in processing and reviewing our manuscript and their encouragements.



Vous devez vous connecter pour continuer.
S'authentifier