1 Introduction
Telomeres are special nucleoprotein complexes at the ends of eukaryotic chromosomes that can switch stochastically between two states: capped and uncapped [1]. Capping is functionally defined as preserving the physical integrity of the telomere. Griffith and de Lange have shown that the 3′ single stranded TTAGGG overhang is tucked back inside the double stranded TTAGGG/CCCTAA telomere DNA, resulting in a large t-loop that is stabilized by an intratelomeric D-loop [2]. A complex of proteins including TRF1 and TRF2 is involved in the stabilization of the telomeric structure [3]. This t-loop/D-loop model could explain how TRF2 protects DNA from the enzymatic attack and end-to-end fusion [4]. However, the molecular mechanism of such t-loop/D-loop is speculative, especially the processes that involve TRF2 [5]. The structures of the TRF proteins themselves and in complex with double stranded DNA have been partially solved by X-ray crystallography [6,7] and NMR [8–10].
In this report, we describe the structure of the Myb-TRF2 and its binding with G-rich strand in random coil conformation or/and in quadruplexes. In order to compare the affinity of Myb-TRF2 with the different components of the t-loop, we also present two repeats telomeric DNA duplex structure itself and in complex with Myb-TRF2.
2 Material and methods
The oligonucleotides d[G(TTAGGG)2T] and d[A(CCCTAA)2C] were purchased from Eurogentec and were passed through Chelex 100 in order to remove the paramagnetic ions.
The proteins were prepared and purified in our laboratory (M.-J. Giraud-Panis, unpublished results).
2.1 NMR experiments
The free oligonucleotides were lyophilized and dissolved in D2O or in H2O/D2O 9:1 (v/v) argon degassed solvent. The protein Myb + 7-TRF2, containing seven N-terminal residues that do not belong to the Myb-like domain, and the protein Myb-TRF2 were used for NMR experiments. These proteins were dissolved in a 50 mM NaCl, 2 mM sodium phosphate buffer (pH 5.3) to which NaN3 was added in order to prevent microbial growth. The NMR samples were degassed then sealed under argon. The DNA duplex titration was made by adding amounts of protein (2.3 mM) dissolved in a 50 mM NaCl, 2 mM sodium phosphate buffer (pH 6.0) to the oligonucleotide duplex (1.25 mM in double strand) dissolved in a 10 mM NaCl buffer (pH 6.0).
NMR experiments were carried out on a VARIAN UNITY-INOVA spectrometer operating at 14.08 Tesla and equipped with a z-axis pulsed field gradient. DQF-COSY, TOCSY with MLEV17 and isotropic period of 40 and 70 ms, and NOESY spectra with mixing time of 50, 100, 120 and 150 ms were recorded at 30 °C. Chemical shifts were referenced to DSS. Sequence-specific 1H NMR assignments were obtained following the conventional strategy [11,12]. Spin systems of the individual amino acids were identified in a D2O solution at 30 °C using DQF-COSY, TOCSY and NOESY spectra. Corresponding spectra in 90% H2O, 10% D2O solution were used to assign the exchangeable protons.
2.2 Structure calculations
The cross-peak intensities on the NOESY spectrum recorded with a 100 ms mixing time were integrated and partially assigned within NMRView [13]. The Myb + 7-TRF2 related NOEs were used as input to ARIA 1.1 [14] implemented in CNS 1.1 [15]. The calculations were performed starting from 150 random templates, using the standard parameters of ARIA (Ambiguous Restraints for Iterative Assignment). ARIA is a method combining an iterative NOE interpretation scheme with a dynamical assignment of ambiguous NOE cross-peaks treated as the sum of contributions from all possible assignments. A large number of NOEs were calibrated and assigned automatically during the structure calculation by ARIA. The procedure of assignment/refinement was repeated iteratively. At each step, the new assignments proposed by ARIA were carefully checked manually and introduced (or not) into the subsequent run. Rejected restraints and residual NOE violations were analyzed, and the assignments were corrected if needed. In the final iteration, the 100 structures with the lowest energy were further refined by molecular dynamics calculations in explicit solvent to remove artefacts [16]. Structures were visualized and aligned with MOLMOL 2.6 [17] and analyzed with PROMOTIF [11], PROCHECK 3.5 [18], and PROCHECK-NMR software programs [19].
3 NMR solution structure of Myb-TRF2
3.1 Structure determination
The protein used for the NMR experiments is described in Fig. 1A and corresponds to Myb + 7-TRF2, the N-terminal seven residues (EDSTTNI) are located upstream the homology domain and the four residues present at the N-terminus do not belong to TRF2. The complete assignment of the proton NMR resonance was obtained from 1H DQF-COSY, TOCSY and NOESY (150 ms) spectra, except for the first residue Gly1, some NH2 resonances of arginine and NH3+ resonances of lysine residues. High field shifted Hα chemical shifts and characteristic Hα(i)–NH(i + 3) and Hα(i)–NH(i + 4) connectivities on the NOESY map provided evidence for the presence of three α-helices involving residues 19–31, 37–43, and 51–64.
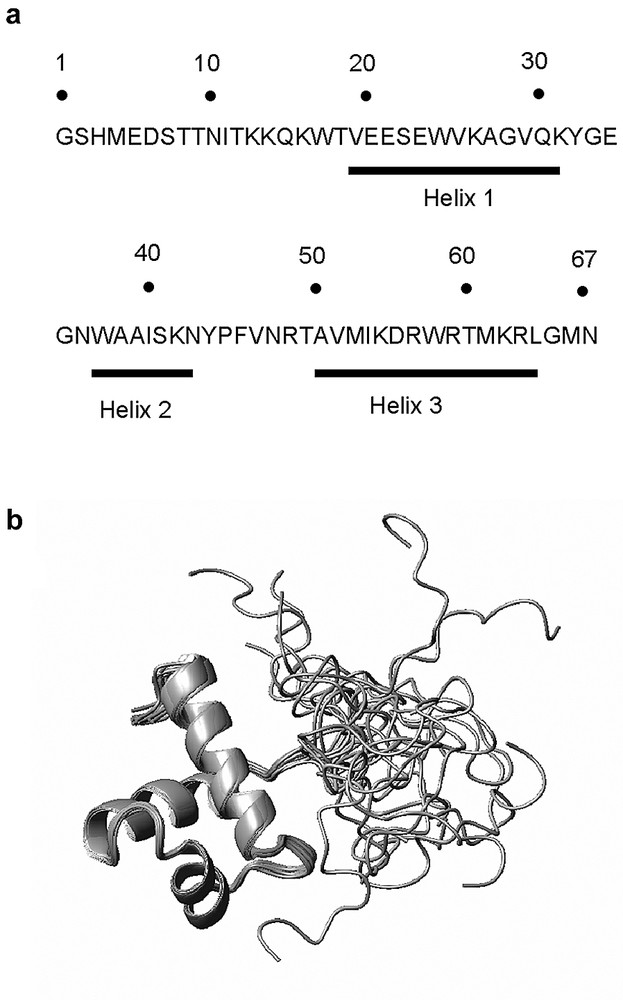
A). Sequence of Myb 438–500; the first four residues do not belong to TRF2. B). An ensemble of 20 refined structures of the DNA binding domain of TRF2 obtained by superimposition of the three helices.
A set of 150 random structures was calculated using the standard ARIA protocol with an ensemble of 444 NOE restraints derived from unequivocal NOEs. After eight simulated annealing iterations of ARIA, all NOE cross-peaks of the 100-ms NOESY map were assigned and 100 structures were refined by molecular dynamics with an explicit solvent environment. Finally, the 20 structures with the lowest potential energies and showing no experimental distance violation larger than 0.3 Å were selected for analysis. The final data file comprising 1754 distance restraints (26 restraints per residue, on average) allowed production of structures with a good convergence. The motif containing the three helices and the two turns (residues 18–67) was well defined with a RMSD of 0.43 ± 0.10 Å on the backbone atoms. Conversely, a global RMSD of 5.87 ± 1.49 Å reflected the lack of structural organization of the N-terminal part. Analysis of the ϕ, ψ angles for the 20 refined structures of Myb-TRF2 indicates that most of the backbone torsion angles (95.5%) lies within the most favored and additional allowed regions of the Ramachandran plot. The residues in the disallowed region (4.5%) were located in turn-1 (Gly33 and Gly35) and in turn-2 (Asn48).
The coordinates of the 20 structures have been deposited with the RCSB Protein Data Bank as entry 1XG1.
3.2 Structural analysis and comparison with Myb-TRF1
The NMR data discussed below correspond to Myb + 7-TRF2. A study of the domain lacking these seven residues was also undertaken. The NOESY spectra of both proteins showed almost exact superposition of the connectivities for the protons of the residues common to the two proteins (data not shown). This result is in agreement with the fact that no NOESY connectivity was observed between the seven first residues and other residues in the sequence. Fig. 1B shows the ensemble of the 20 calculated structures. One can notice the high definition of the structure from residue 18–67 and the mobility found in the N-terminal arm. The folding of the protein can explain some unusual shifts observed for several protons in the NOESY spectra. The amide proton of Val47 appears to be in the plane of the cycle of Trp24, in agreement with the observed downfield shift of its resonance at 9.85 ppm [Hζ3(Trp24)–NH(Val47) = 3.0 Å]. The amide proton of Gly35 and Cα proton of Lys55 are located on both sides and near to the cycles of the tryptophan Trp37, in accordance with the strong up field shift observed for their resonance: 5.45 and 1.78 ppm respectively. One Hα of Gly28 due to its proximity with the Tyr32 and Tyr44 rings is low field shifted at 4.62 ppm. The hydroxylic protons of Thr18 and Thr60 are hydrogen bonded to the carboxylate group of Glu21 and to the carbonyl group of Arg57 respectively, this explains that their resonance was observed and located at 5.66 and 5.12 ppm.
The overall structure of the folded domain is very similar to that of TRF1 with the presence of three α-helices (residues 19–31, 37–43 and 51–64) linked by two turns (residues 32–36 and 44–50). A hydrophobic core is formed by the residues Trp17, Val25, Val29, Trp37, Ile40, Phe46, Ala51, Ile54, Trp58, and Met61. Most of these residues are conserved between Myb-TRF1 and Myb-TRF2 (Val25 and Ile54 in Myb-TRF2 instead of Leucines in Myb-TRF1) and in both structures they are involved in maintaining the three helices. The first helix resembles that of Myb-TRF1 in length and relative orientation but presents a different distribution of the residues along the surface. In Myb-TRF1, two faces are clearly defined on the structure, one hydrophobic and one charged. In Myb-TRF2 residues of a different nature are mixed along the helix.
The first turn, classified as a β-turn IV, is stabilized by two hydrogen bonds between NH(Asn36) and CO(Gly35), and between NH(Trp37) and CO(Gly35). Helices 2 and 3 form the variant of the helix–turn–helix motif responsible for most of the interactions between Myb-TRF1 and DNA [8]. Although being very similar to Myb-TRF1, helix 2 has a slightly different position in Myb-TRF2 due to the presence of Pro45, which induces the formation of a γ-turn (44–46). This second turn is stabilized by the interaction of Phe46 ring (a conserved residue) with the side chain of a hydrophobic residue in each helix: Val25 (a leucine in Myb-TRF1), Ile40 (conserved) and Ile54 (another leucine in Myb-TRF1). Two hydrogen bonds between HN(Phe46) and CO(Tyr44), and between NεH(Arg49) and CO(Val47) participate also to the stability of this structural element. Following the γ-turn (44–46) that is missing in Myb-TRF1, the end of the turn (46–49) recovers a similar position between both domains as a classical β-turn IV. Helix 3 is undistinguishable from that of Myb-TRF1 owing to the remarkable homology between both sequences (11 residues are conserved in the 14 forming the helix). Could the differences that we observed between Myb-TRF2 and Myb-TRF1, be responsible for differences in DNA binding behavior? The most likely answer to that question is no. Indeed, the overall shape of the molecule is uncannily similar and the small differences observed occur in regions of the molecule that are not involved in DNA interactions in Myb-TRF1. Finally the vast majority of the residues that are implicated in DNA binding are conserved between the two proteins and in both cases are exposed to the solvent (Trp17, Arg49, Met53, Lys55, Asp56, Arg57, Arg59, Thr60 and Arg63).
3.3 Single stranded telomeric human DNA structure
It is known that the C-rich human telomeric strand d[(CCCTAA)3CCC] adopts an i-motif structure at acidic pH [20] and that d[AGGG(TTAGGG)3] folds on itself as a G tetrad [21].
At DNA concentrations ≤ 0.2 mM and in absence of salt, the 1H NMR spectra of d[G(TTAGGG)2T] were almost independent of the temperature between 5 and 70 °C, demonstrating a random coil conformation in agreement with the absence of imino protons resonances. Adding KCl led to the appearance of several sharp imino resonance in the range of 10–11.5 ppm. These resonances were assigned to quadruplexe structures since it has been shown that the four-repeats G-rich strand folded in a single strand with quartets of guanine [21] and that the two repeats human telomeric DNA showed parallel or antiparallel quadruplexes in presence of salt and after substitution of thymines in uracil or bromouracil [22].
The NMR spectra of 10−4–10−3 M d[A(CCCTAA)2C] showed sharp proton resonance without resonance in the range of 10–16 ppm, even at pH 5.0 demonstrating a random coil single stranded conformation.
3.4 Titration of the two repeats G-rich telomeric strand with the protein Myb-TRF2
Adding successive amounts of Myb-TRF2 (0.0–0.2 mM) to d[G(TTAGGG)2T] (0.2 mM) in random coil conformation led to a broadening of the resonance lines and slight shifts of the oligonucleotide resonances. It is worth noting that inter-residue NOE connectivities (H6/8–H1′, –H2″, –H6/8, –H5, and –CH3) were well distinguishable on the 150 ms NOESY spectrum at 25 °C, whereas they were absent with the single strand. This corresponds to the fact that the random coil conformation exhibited large internal dynamics, which led to an effective correlation time so that ω τ ≈1.1 and reduced dramatically the NOE efficiency. The presence of well-definite NOE connectivities corresponding to the increase of the effective correlation time strongly suggested the binding of Myb-TRF2 to the single-stranded d[G(TTAGGG)2T] in random coil conformation.
Adding Myb-TRF2 (0.2 mM) to the quadruplex form (0.2 mM) in presence of KCl (50 mM) led to several changes in the 1-D proton spectrum and to the appearance of several intermolecular NOE connectivities between the quadruplex and the protein. It is well known that unspecific binding of protein to DNA leads to the formation of several types of complexes, each of them owing different contact points with low populations, generating undetectable NOEs. On the contrary, specific interaction only produces one type of complex where NOEs observation is expected. Such situations were observed in NOESY spectra of specific and unspecific complexes between the Lac Repressor headpiece and DNA for example [23]. Consequently, presence of intermolecular NOEs is indicative of specific binding. Our data strongly suggest a tight binding of Myb-TRF2 with the quadruplex.
3.5 Titration of the telomeric DNA duplex by the protein Myb-TRF2
The 1:1 mixing of the strands d[G(TTAGGG)2T] and d[A(CCCTAA)2C] in absence of salts, showed NMR spectra with exchangeable and non-exchangeable proton resonances corresponding to a right-handed duplex conformation, even at pH 5.5, despite the possible folding of the separate strands. The N1H imino protons of the guanines were found in the range of 12–12.5 ppm and the N3H imino protons of the thymines were located in the range of 13–14 ppm. No imino resonances corresponding to quadruplexes species or protonated cytosines were observed. All the exchangeable and non-exchangeable proton resonances were sequentially assigned to a B-type right-handed helix except the 5′ and 5″ ones that were found to overlap.
Adding successive amounts of protein to a 0.25 mM solution of DNA at 20 °C led to the increase of new set of bound imino protons resonances and to a decrease of the free-form resonances until the ratio 1/1 between protein and DNA was reached (Fig. 2). These spectra gave evidence of a slow exchange process on the NMR time scale between the free form and the bound form of the DNA, suggesting a tight binding of the protein to the telomeric DNA.
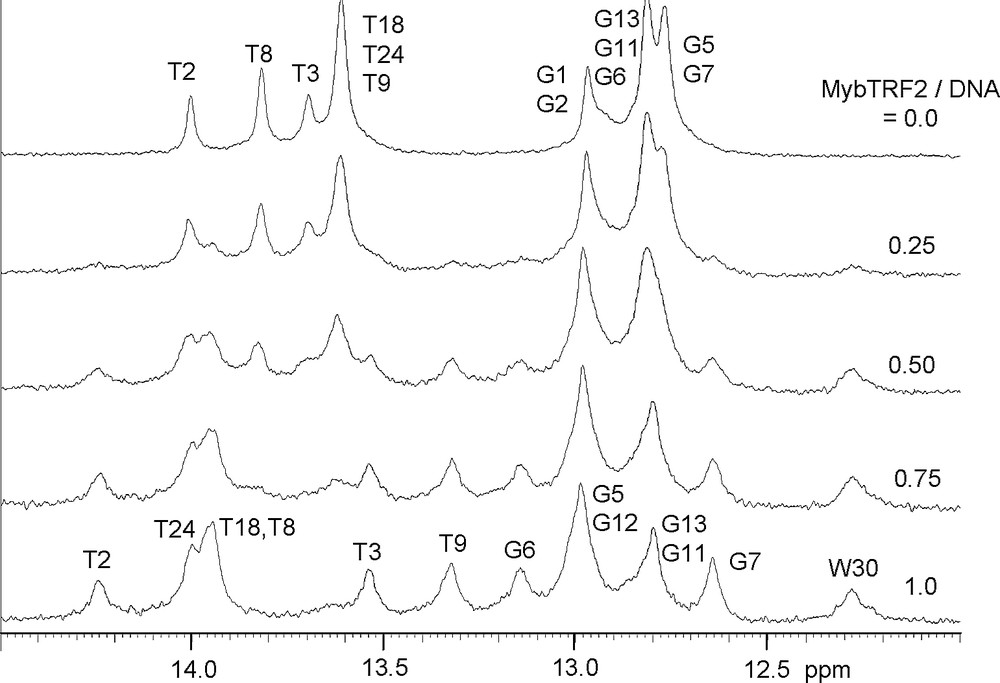
Imino proton resonance spectra of two repeats duplex in the presence of increasing amounts of Myb-TRF2 showing slow chemical exchange.
It can be pointed out that the binding of Myb-TRF2 without the seven supplementary residues led to very similar spectra with the same imino proton resonances in slow chemical exchange. We conclude that these seven supplementary amino acids did not belong to the specific protein fixation domain.
3.6 Contribution of individual base pairs to the sequence-specific binding
The binding specificity of Myb-TRF2 with the two repeats telomeric DNA was tested by running NOESY spectra at 50, 150 and 300 ms mixing time (Fig. 3). Several intermolecular NOESY connectivities were observed, indicating a specific interaction. Although a complete assignment of these connectivities requires the use of NOESY experiments with half filter on a mixture of labeled and unlabeled molecule, we were able to assign some of them: NH(W58) with H8(A22), H3′(A22), H1′(A22) and H1′(C21); Hβ(W58) with H8(A22); CH3(V19) with H5(C19) and H5(C20); CH3(T60) with H1′(A22) and H3′(A23).
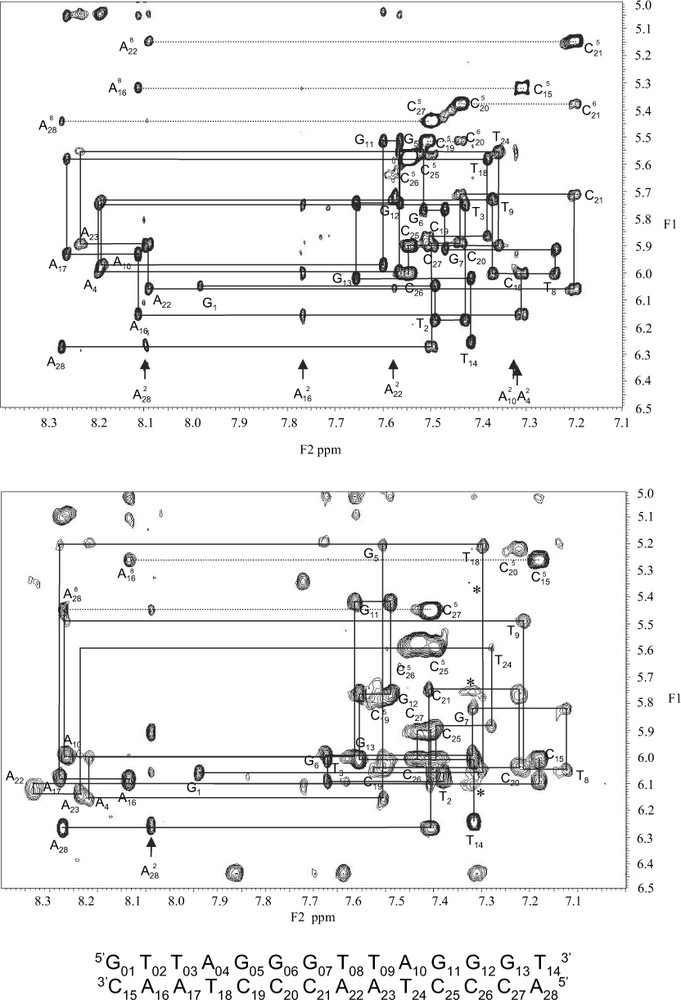
Expansion of the H6/8-H1′ NOE connectivities of the duplex free (top) and complexes to Myb-TRF2 (bottom).
Intermolecular cross-peaks corresponding to NH(W58) (7.32 ppm) with H1′(C21), H1′(A22), and H3′(A22) (5.35 ppm) are showed by a star.
In order to estimate the contribution of each base pair in the telomeric binding site, the assignment of the imino, aromatic and H1′ resonances of the DNA was made for the free and bound DNA. The lack of protein resonances in the range of 12–15 pomp, except the NεH of the tryptophan 37 at 12.27 ppm, and the small number of NOESY protein–protein connectivities between non-exchangeable protein protons in the range of 7–9 ppm allow us to assign without difficulty almost all the imino, aromatic and H1′ resonances of the bound DNA. Important chemical shift variations were observed for these protons located in the minor groove and in the major groove of the DNA. The most important variations were observed for the paired nucleotides T2-A16 until A10-T24. The imino proton resonances of the residues T2, G5, G6, T8, T14, T18 and T24 were downfield shifted, indicating a strengthening of their hydrogen bonds. The imino resonance of T9 were upfield shifted as well as the H2 proton of its paired adenine A23, suggesting an increase of the stacking of the base pair T9-A23 with its surrounding T8-A22 and A10-T24 base pairs. It is worth noting that the H2 resonance of the base pair T3-A17 did not exhibit such a trend. These chemical shift variations indicated that the binding domain of Myb-TRF2 overlapped parts of each of the two motifs TTAGGG, in agreement with the formation of a 1–1 complex. The same observation can be made with the H1′ protons located in the minor groove like the H2 protons. Several important shifts were also observed for some aromatic resonances (H5, H6 and H8) corresponding to protons of the residues T3, G6, C19, C20, C21 and A22 located in the major groove of the double helix. The NεH of the tryptophan 37 was downfield shifted by 1.2 ppm upon complexation suggesting the formation of a hydrogen bond or of an electrostatic bond. All these data are in agreement with the crystallographic structure reported for the complex with TRF2 [7].
4 Conclusion
We demonstrated here, that Myb-TRF2 binds to the duplex, to the random coil G-rich strand and to the quadruplex of the human telomeric DNA, contrary to already reported results [24]. The great similarity of the structures of Myb-TRF1 and Myb-TRF2 as well as their NMR features suggests a similar binding mode to the double stranded telomere. It is therefore clear that TRF2 functional singularity cannot originate from the DNA binding domain per se. Nevertheless these data are important to understand the formation of the t-loop since it requires the binding of TRF2 and the coexistence of single strand, double strand and presumably of quadruplex [25].


