1 Introduction
The Son of sevenless (Sos) nucleotide exchange factor is a protein of 1333 amino acids known to be crucial in tyrosine kinase receptor-mediated signaling processes. Indeed, it interacts with two ∼60 residues Src homology 3 (SH3) domains of the membrane Growing Factor Receptor-bound-2 protein (Grb2) through a short proline-rich region, VPPPVPPRRR (V10R, residues 1149 to 1158). This interaction induces the conversion of Ras-GTP into Ras-GDP, followed by the activation of both MEK and ERK proteins, which finally leads to cell response (multiplication, differentiation).
Numerous works have focussed on the development of V10R mimicking inhibitors, which are able to disrupt the upstream Grb2/Sos complex [1]. Among the most promising derivatives, dimeric V10R ligands [3], peptoid analogues [4], or peptides incorporating Cβ-substituted proline [1] have been described. Structure of the V10R/Grb2 complex has been determined by NMR [2] and served as a rationale for new inhibitors design. Bound peptide was found to adopt a PPII conformation (ϕ ∼ −75°; ψ ∼ 145°) from residue Pro2 to Pro7. However, little is known on the structure of the free V10R peptide in solution. In this regard, the presence of five prolines may be associated with conformational exchange and thus could reduce the concentration of the bioactive conformation of V10R. The present work aims first to explore which conformations are present in solution and second to calculate the structure of the major conformer on the basis of NMR constraints.
2 Experimental section
2.1 Peptide synthesis
Automatic peptide synthesis was achieved as described elsewhere [1,5]. The H–VPPPVPPRRR–OH peptide (V10R) was obtained from a Boc–Arg(Tos)–OH preloaded PAM resin (0.1 mmol): RP-HPLC, tR = 10.3 min (30 min gradient from 9 to 48% CH3CN). MALDI-Tof (M + H+) 1170.7 (calcd 1169.7). The N-acetylated and C-carboxamide V10R (Ac–V10R–NH2) peptide was obtained from a MBHA resin. After removal of the last Boc-protecting group, the amino terminal was acetylated by an excess of acetic anhydride. RP-HPLC, tR = 15.9 min (30 min gradient 6 to 30% CH3CN). MALDI-Tof (M + H+) 1211.9 (calcd 1210.7).
2.2 Circular dichroism
CD spectra were acquired on a Jobin Yvon CD6 dichroism spectropolarimeter at 5 °C using a 1-mm pathlength quartz cell. Spectra were recorded from 190 to 260 nm and were averaged over four scans after baseline correction. Samples containing 2.0–7.4 M guanidinium hydrochloride (Merck) were obtained by adequate additions of 8.0 M concentrated solution. Samples containing 2.0–5.5 M calcium chloride (Sigma–Aldrich) were obtained by adequate additions of 6.0 M concentrated solution. For each sample, final V10R peptide concentration was fixed to 300 μM in a 2 mM phosphate buffer, pH 7.
2.3 NMR spectroscopy
NMR samples were prepared by dissolving lyophilized peptides (V10R or Ac–V10R–NH2) to final concentrations of 2 mM in 550 μL phosphate buffer (20 mM, pH 6.7 in 10% or 100% D2O). Sodium 2,2-dimethyl-2-silapentane-5-sulfonate-3,4,5-d6 at 0.5 mM was used as an internal reference for 1H chemical shift calibration. NMR experiments were recorded at 25 °C on a DMX Bruker Avance spectrometer operating at a 1H frequency of 500 MHz and equipped with a room-temperature, triple-resonance, z-axis pulsed-field-gradient probehead, optimized for 1H detection. Proton assignments were obtained from the analysis of 2D total correlation spectroscopy (TOCSY) experiments [6] using a 65 ms DIPSI-2 mixing time [7], 2D waterflipback nuclear Overhauser enhancement spectroscopy (NOESY) [8] and 2D rotating frame Overhauser effect spectroscopy (ROESY) [9] experiments (250 ms mixing time). Solvent suppression was achieved with a WATERGATE sequence [10]. Two-dimensional experiments were typically collected as 512 (t1) and 2048 (t2) time-domain matrices over a spectral width of 11 ppm, with 32 scans per t1 increment. Carbon assignment was deduced from proton assignment using heteronuclear 2D 1H–13C HSQC spectra [11].
Data were processed with NMRPipe software running on a SGI O2 workstation [12]. Shifted sine-bell window functions were applied in both indirect and direct detected dimensions and extensive zero filling prior to Fourier transformation was used to yield high digital resolution. Spectra were analyzed using CARA software [13].
2.4 Structure calculation
Simulated annealing (SA) calculations were carried out with the program DYNAMO 2.1 [14] and consisted of three stages. The first stage comprised of an initialization period of 1000 steps (3 fs long) of molecular dynamics at 4000 K and very tight temperature control. In the second stage, the coordinates were allowed to cook at 4000 K with lose temperature control for 4000 steps. In the final stage, the structure was annealed by slowly reducing the temperature from 4000 to 0 K over the space of 20,000 molecular dynamic steps (3 fs long). In the first two stages, force constants were set as follows: 1000 kcal mol−1 Å−2, 250 kcal mol−1 rad−2, 50 kcal mol−1 rad−2, 10 kcal mol−1 rad−2, 2 kcal mol−1 Å−2 for bonds, angles, impropers, torsions and NOE constraints, respectively. No Van der Waals interactions were operative. During the final stage, bond force constant was maintained at 1000 kcal mol−1 Å−2, torsions were ramped from 10 to 200 kcal mol−1 rad−2, both angles and impropers to 500 kcal mol−1 rad−2, NOE term was increased exponentially from 2 to 30 kcal mol−1 Å−2. The Van der Waals interactions and J-coupling restraints were slowly introduced during cooling to reach the final values of 4 kcal mol−1 Å−2 and 1 kcal mol−1 rad−2, respectively. Six ϕ- and five ψ-torsions restraints have been predicted with good accuracy (ca. ±10°) using the program TALOS [15] and have been included in structure calculation. Other NMR constraints consisted of 16 vicinal J-coupling constants measured from 1D and/or 2D spectra and 62 NOE derived from NOESY and ROESY experiments. Hundred structures have been calculated starting from an extended fold. Typical final energies (kcal mol−1) were (<0.2); (3.8 ± 0.3); (4.5 ± 0.5); (3.2 ± 0.4); (22.7 ± 1.5); (4.7 ± 0.5) and (14.9 ± 1.3) for torsion, J-coupling, bond, impropers, angle, NOE and Van der Waals force constants, respectively.
3 Peptide bond conformation
Due to its partial double bond character, the planar peptide bond may adopt either a cis or a trans conformation (ω = 0° or 180°, respectively). However, in peptides and proteins, the cis population remains marginal because unfavourable steric repulsions occur between the two sequential CHα methyne. It should be stressed that in the case of proline residues, the presence of the N-attached CHδ group destabilizes also the trans conformation, reducing therefore the energy difference between the two conformers. A statistical analysis based on a non-redundant set of 571 proteins showed that the fraction of cis peptide bonds was as low as 0.03% for Xaa–nonPro amide bonds, but reached 5.20% for Xaa–Pro bonds [16]. It was also reported that for peptides or proteins in solution, one given Xaa–Pro peptide bond could adopt both conformations, the two conformers being in slow exchange [17,18].
3.1 Relative stability of conformers
Since V10R contains five prolines, 25 different conformers may be in slow exchange due to the cis–trans proline isomerisation. Then, a preliminary 1D NMR analysis has been performed to detect the number of V10R conformers present in solution. Four signals are expected per conformer, the 10 residues peptide containing five proline and the first residue amine protons being not visible in the spectrum because of fast exchange with water. Surprisingly, the 1H amide region contained only few peaks, including four strong and four weak doublets (Fig. 1). One can directly conclude from this observation that among the possible 32 conformers only two were present in solution (conformers A and B). Moreover, signal integration gave a ratio of 90:10 for conformers A and B, respectively.
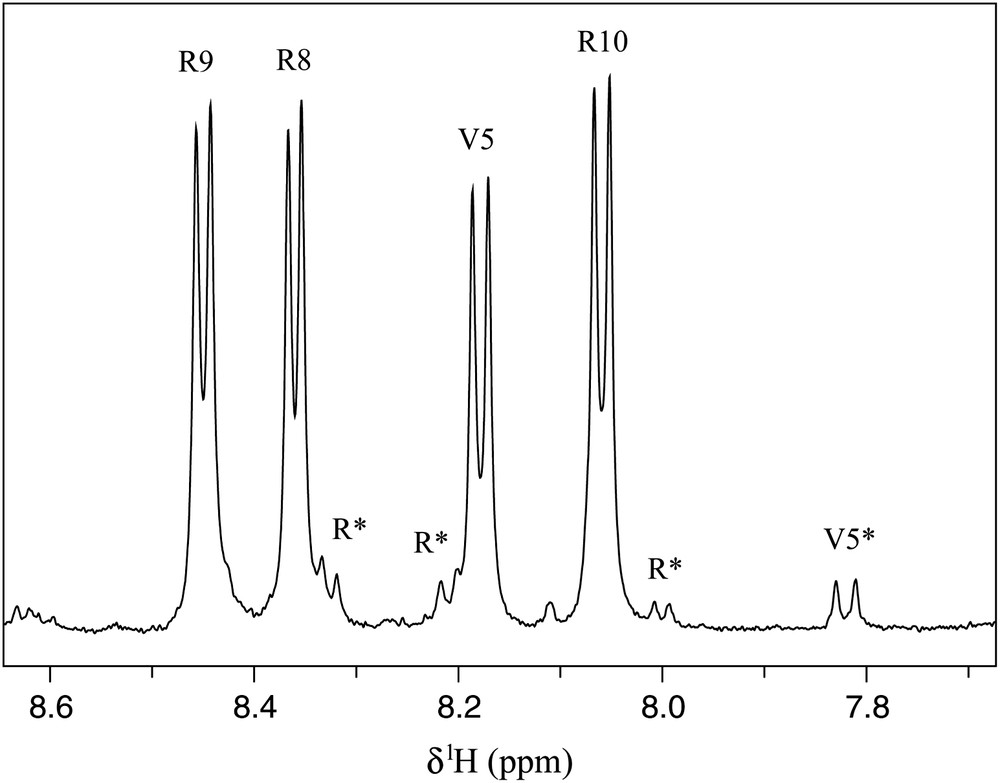
Amide region of the 1H NMR spectrum of V10R (2 mM) recorded at 500 MHz in 20 mM phosphate buffer pH 6.7, H2O:D2O (90:10), 25 °C. Assignments of conformers A and B have been reported on the spectrum using one-letter amino acid code. Asterisks refer to conformer B resonances.
The same result was deduced from close inspection of 2D 13C–1H HSQC spectra. In addition to two strong correlation peaks in the Cα–Hα valine region assigned to V1 and V5, one minor correlation (9%) was observed for B, suggesting that one of the two valine residues was sensitive to a slow exchange between two V10R conformers (Fig. 2a). The presence of this exchange was also confirmed in the Cγ–Hγ methyl region where two weak additional methyl correlations were observed (Fig. 2b). It is noteworthy that one of the two valine residues was not influenced by the conformational change of V10R, probably because proline isomerisation took place far in the sequence and did not induce chemical shifts perturbation.

Valine 1H–13C correlation peaks of V10R (a, b) and Ac–V10R–NH2 (c, d). HSQC spectra were recorded at 500 MHz, 25 °C, using samples in 20 mM phosphate buffer pH 6.7, D2O 100%. Assignments of conformers A and B have been reported on the spectra using one-letter amino acid code. Asterisks refer to conformer B resonances.
Stability of the two conformers was also assessed as a function of temperature. Vicinal were very stable when raising the temperature from 5 to 65 °C (ca. < 0.01 Hz/°C, data not shown). Moreover, the number of conformers did not increase at high temperature as judged by analysis of proton 1D or 2D 13C–1H spectra, and the ratio between the conformers A and B being also about 90:10 at 65 °C. No exchange peak between the two V10R forms was detected in NOESY spectra, even using long mixing time delays at high temperature [18]. Altogether, our data suggested that whatever the temperature (5–65 °C), two V10R conformers in a stable ratio of 91:9 were interconverting in the slow exchange regime.
3.2 Conformers properties
Complete and partial 1H and 13C assignments have been obtained from homo- and heteronuclear 2D spectra for conformers A and B, respectively. Proline 13Cβ and 13Cγ chemical shifts give direct information about Xaa–Pro peptide bond conformations [19]. Indeed, the Cβ–Cγ correlation pattern of conformer A (Fig. 3) revealed that Pro2, Pro3, Pro4, Pro6 and Pro7 belong unambiguously to the trans region plot. This latter conformation was confirmed by NOESY spectra since strong HαXaai−1–HδProi NOE were observed for these residues. Interestingly, the Cβ–Cγ correlation plot of conformer B placed its Pro6 residue in the cis region. This peptide bond conformation was further supported by the NOESY spectra where a strong HαVal5–HαPro6 cross peak was observed (Fig. 4). Thus, the five prolines of conformer A were found in the trans conformation, a feature compatible with a PPII fold, whereas the Pro6 residue of conformer B was in the cis form. This provides explanation why the immediately preceding Val5 experienced different chemical shifts for each conformer. In contrast, chemical shifts of other residues appeared unchanged or very close for conformers A and B, suggesting that Pro6 conformational state is the only structural difference between A and B.
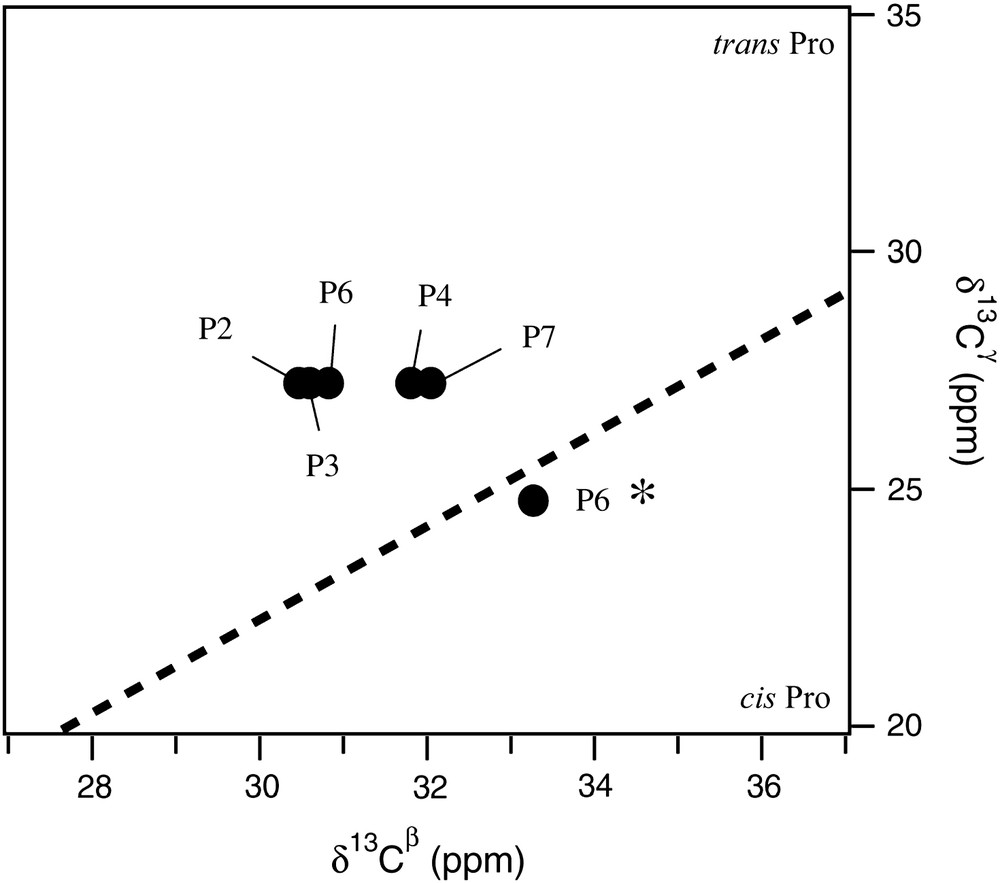
Cγ–Cβ correlation plot of proline residues. Proline assignment of conformer A has been reported on the spectra using one-letter amino acid code. P6* refers to Pro6 of conformer B.
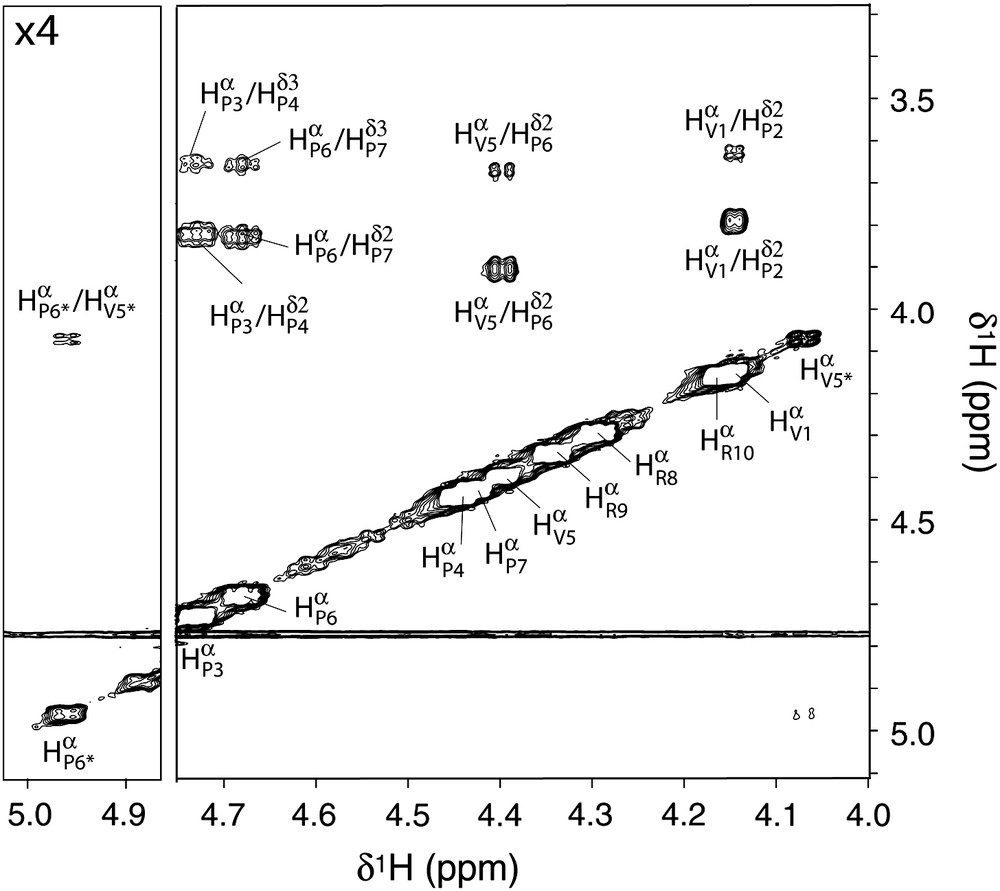
Hα region of the NOESY spectrum, recorded at 500 MHz, 25 °C, using samples in 20 mM phosphate buffer pH 6.7, D2O 100%. Assignments of conformers A and B have been reported on the spectra using one-letter amino acid code. Asterisks refer to conformer B resonances.
3.3 Influence of the N-termination on peptide structure
The five V10R prolines can be classified into three groups: (i) prolines preceded by a non-prolyl residue (Pro2 and Pro6), (ii) prolines followed by a non-prolyl residue (Pro4 and Pro7) and (iii) prolines surrounded by two prolines (Pro3). One can observe that Pro6, the only proline experiencing cis–trans isomerisation, belongs to the group (i). However, the Val1–Pro2 peptide bond was >99% trans, as estimated from 13C–1H spectra (Fig. 2a and b). To assess whether the N-termination of Val1 could influence Pro2 isomerisation, Ac–V10R–NH2 was synthesised. Interestingly, the presence of the N-terminal acetyl group restored the ability of Pro2 to undergo the two conformational states. Indeed, secondary signals were observed for Val1 of Ac–V10R–NH2 in both the Cα–Hα and Cγ–Hγ regions (Fig. 2c and d, respectively). Precise integration of strong and weak Cα–Hα, Cγ1–Hγ1 and Cγ2–Hγ2 correlation peaks gave 88% of trans conformation for the Val1–Pro2 peptide bond in Ac–V10R–NH2, whereas the trans Val5–Pro6 peptide bond remained unchanged at 91%.
4 Circular dichroism experiments
The PPII content of a peptide may be quantified using far-UV circular dichroism experiments. Indeed, previous works showed that a characteristic positive band at 228 nm was observed for pure PPII peptides [20]. Moreover, guanidinium hydrochloride has been shown to stabilize PPII conformation [20], whereas calcium chloride destabilizes PPII conformation [21]. Thus, V10R samples containing increasing amounts of guanidinium hydrochloride or calcium chloride have been analyzed by far-UV CD (Fig. 5). Only a residual ellipticity increase was observed at 228 nm when raising guanidinium hydrochloride concentration from 7.0 to 7.4 M, suggesting that the maximum 100% PPII was almost reached. Similarly, the minimum ellipticity corresponding to 0% PPII was obtained with 5.0 M calcium chloride since no spectral change occurred between 5.0 and 5.5 M. These results allowed us to determine at 228 nm an ellipticity dynamic range from 0 to 100% PPII of 760 deg cm2 dmol−1. According to this statement, a ∼60% ratio of PPII helix has been estimated for free V10R in solution, in good accordance with results obtained on similar peptides [20].
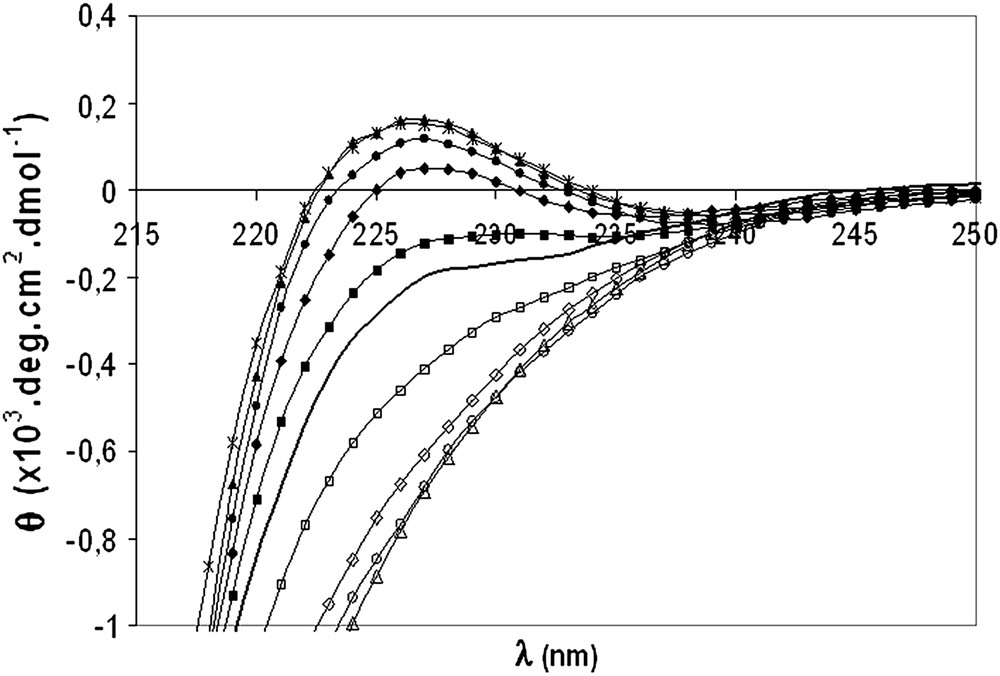
CD spectra of V10R in 2 mM phosphate buffer, pH 7 (bold line) and in the presence of 2.0 M (■), 4.0 M (♦), 6.0 M (●), 7.0 M (▴), 7.4 M () guanidinium hydrochloride or in 2.0 M (□), 4.0 M (♢), 5.0 M (○) and 5.5 M (▵) calcium chloride.
5 Structure of conformer A
5.1 Structure calculation
Structure determination of conformer A has been carried out by NMR spectroscopy. Constraints consisted of a total of 62 NOE, 11 torsions restraints, 4 and 12 vicinal couplings. Among the 100 calculated structures, the best 10 are presented in Fig. 6. The first Val1–Pro7 segment adopted a left-handed helix conformation with three residues per turn. Proline torsion angles were closely restricted to the canonical PPII ϕ and ψ values (−67 ± 7° and 150 ± 12°, respectively). Interestingly, proline residues belonging to group (ii) (Pro4 and Pro7) had a slight 5–10° weaker ψ angle than those followed by prolines. Since Val1 (ψ ∼ 148°) and Val5 (ϕ ∼ −85° and ψ ∼ 132°) were also located in the PPII region of the Ramachandran plot, the overall fold of the Val1–Pro7 segment was a regular PPII helix, characterised by a very low RMSD (RMSD = 0.35 Å for backbone atoms and RMSD = 1.67 Å for all atoms taken from the 10 best structures). In contrast, the three C-terminal Arg residues were poorly structured, as a result of the lack of NOE constraints.
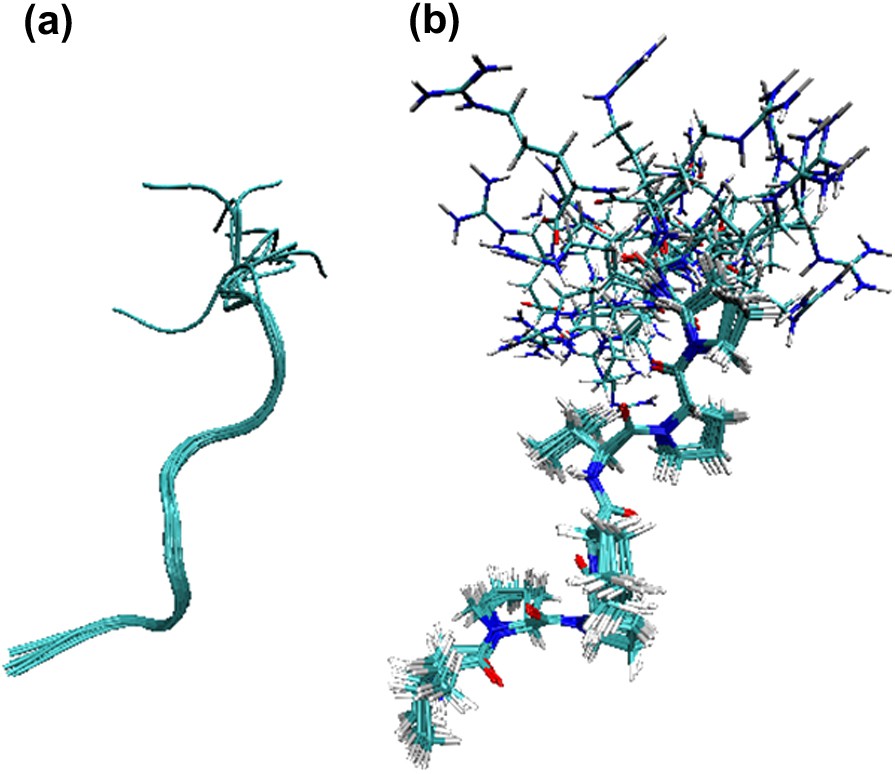
Superposition of the 10 best V10R structures (conformer A). (a) Backbone RMSDs were 0.35 Å for residues Val1–Pro 7 and 1.25 Å for the entire peptide, (b) RMSDs for all atoms were 1.67 Å for residues Val1–Pro 7 and 3.13 Å for the entire peptide.
5.2 Comparison of the free and Grb2-bound structures
Structure calculations unambiguously led to an extended PPII structure for conformer A, which accounts for 91% in solution. Alignment with the structure obtained for the Grb2-bound peptide [2] gave a small RMSD value of 0.96 Å for backbone atoms and a RMSD of 1.65 Å for all atoms belonging to residues comprised between Val1 and Pro7. Close analysis of dihedral angles of this region revealed same features for both structures, suggesting that no major conformational change occurs when conformer A of V10R binds to Grb2 within this segment. In contrast, the basic C-terminal region of V10R must be stabilised upon binding since Arg8 directly interacts with an aspartate of the protein (Asp33 and Asp190 for the Grb2 N-terminal and C-terminal SH3 domains, respectively).
6 Conclusion
The structural study by NMR and far-UV circular dichroism of the free V10R peptide is reported. For temperatures ranging from 5 to 65 °C, 1H and 2D 13C–1H NMR spectra revealed the presence of two conformers slowly interconverting in a stable ratio of 91:9. The structure of the main conformer corresponds to a well-defined PPII helix for the Val1–Pro7 segment, and an unstructured three-residue C-terminal tail. Thus, all proline peptide bonds were trans for the conformer A, whereas a cis Val5–Pro6 peptide bond was detected for the minor V10R form (conformer B). Far-UV CD analysis was in agreement with NMR data since ∼60% PPII conformation was found (69% by NMR).
The structural alignment of conformer A and the active Grb2-bound V10R was performed, giving a very low RMSD between both structures. This suggested that conformer A could bind to Grb2 without major conformational rearrangement. In addition, analysis of the Ac–V10R–NH2 structure suggested that the 90:10 ratio for trans–cis peptide bond was a common feature for proline preceded by a N-amide non-proline residue. We previously showed that N-acetylation and C-carboxamidation of V10R gave a 10-fold increase in affinity for Grb2 [1]. However, the present study was indicative of a slight lower active conformation for the free Ac–V10R–NH2 than for V10R, due to 10% Val1–Pro2 peptide bond isomerisation. Thus, it can be concluded that differences in Grb2 affinity are mainly related to electrostatic effects.
Acknowledgements
We are grateful to Dr. Fabienne Burlina for mass spectra analysis and to Vanessa Point for experimental assistance. We are also grateful to Dr. S. Fermandjian and Dr. L. Zargarian (CNRS-UMR 8113, Institut Gustave-Roussy, Villejuif) for circular dichroism experiments.


