1 Introduction
Uric acid and urate salts occur as deposits in urinary calculi [1–5] as well as in gouty arthritis [6]; a detailed knowledge of the crystal structures involved is thus of biological importance [7–10]. Of the compounds of biological interest, only a few are characterized crystallographically: anhydrous uric acid [11], its monohydrated [12] and dihydrated [13] forms, as well as monosodium urate monohydrate [14], ammonium urate from powder diffraction data [15] and calcium urate hexahydrate CaUr2·6H2O [16]. Concerning the potassium derivatives it is difficult to know exactly what is behind the name “(mono)potassium urate” abbreviated sometimes as MPU or KU, said to be hydrated or not, and mentioned in numerous papers including a recent report Ref. [17], since no crystal structure is yet available. Potassium urate, KC5H3N4O3, is number 728 in the famous Index of Powder Diffraction Data for 1000 Chemical Substances [18], which was a seminal work at the origin of the Powder Diffraction File (PDF) database. This compound is still the only potassium urate in the PDF-4 Organics database [19] with the entry number 00-001-0302. Finally, a potassium urate was claimed to be identified in urinary tract calculi [20].
Chemically, the title compound corresponds exactly to the “Quadrurate of potassa” KH(C5H2N4O3), H2(C5H2N4O3) as named by Bence Jones [21], believed to be amorphous, discussed later by Roberts [22,23] who renamed it potassium quadriurate [23]. In this nomenclature, KH(C5H2N4O3) is a biurate, nowadays called (mono)potassium urate, but this old name is still used as a synonym by some chemical suppliers. According to Sir William Roberts [22], the quadriurates (of K, Na, NH4) “exist in nature in the form of the amorphous urate sediment of human urine, and as the essential constituent of the urinary excretion of birds and serpents.” Moreover “the quadr(i)urate is the form, and the only form, in which uric acid exists in normal urine” and “when uric acid makes its appearance therein, such an event is due to secondary changes in the quadr(i)urate.” That theory of a double salt, made up of a molecule of uric acid combined with a molecule of an acid urate, was further examined by Kohler [24] who declared that there was no such body as quadriurate. We present here the first recognized crystalline occurrence of a potassium quadriurate as revealed by Structure Determination by Powder Diffractometry (SDPD) methodologies [25]. Being microcrystalline, its crystalline nature may have escaped attention due to the limited means for microscopical observations during the XIXth century where its granular appearance without any obvious crystalline structure led to the conclusion that it was amorphous, or, as stated by Kohler [24], the samples were really just mixtures of urate and uric acid.
2 Experimental
2.1 Origin of the sample
The calculus used for this study is from a 63 year old man presenting recurrent stones for several years, often expelled spontaneously. There were other calculi in the kidneys and bladder. We received three calculi at the Tenon Hospital, Paris; two were made of anhydrous uric acid. Qualitative chemical analysis and IR spectra similarity led to the conclusion that the third calculus could be an oxypurine potassium salt (no trace of Na, Mg or Ca). Given the difference in composition, and probably of pH necessary for their formation, it is likely that these calculi had different anatomical origins, but we have no way of knowing more. FTIR measurements are displayed in the supplementary material. The presence of an amorphous component cannot be excluded at this stage.
2.2 Synchrotron measurements
X-ray diffraction measurements were conducted on the Cristal beamline of synchrotron SOLEIL (France). The X-ray wavelength of 0.72442 Å selected using a double Si(111) crystal monochromator was determined by recording a diffraction pattern of a standard LaB6 powder (NIST SRM 660b). The sample, introduced in a glass capillary (ø = 0.7 mm), was mounted on a spinner rotating at 5 Hz to improve particle orientation averaging. Data were collected in Debye Scherrer mode using a 21 Si(111) crystals analyzer [26]. With this setup, three high angular resolution diagrams recorded in about two hours were summed up since sample degradation under the X-ray beam was not significant.
3 Structure determination and refinement
Attempts of identification by search-matching using the Bruker EVA [27] software combined with the PDF database [19] failed: no trace of the previously reported Ref. [18] potassium urate (00-001-0302) was found in our sample. Indexing of the synchrotron powder pattern of the potassium quadriurate was realized by using the McMaille software [28], leading to a primitive monoclinic cell. The Figures of Merit (FoMs) were M20 = 115.7 [29], F20 = 518.2 (0.0012, 33) [30], and McM20 = 184.7 [31]. The space group determination was unambiguously leading to P21/c. A quite satisfactory Le Bail fit [32] confirmed the indexing and was used to extract the intensities. Since the structure determination was undertaken without a detailed knowledge of the chemical composition, several oxypurine molecules were tested (uric acid, allantoin, hypoxanthine, and xanthine) and various K: molecule ratios.
The cell volume suggested the possibility of two independent molecules. The structure solution was carried out in direct space by using the ESPOIR software [33], moving in the cell the molecules and the potassium atoms by a Monte Carlo process. A clear best result was obtained for two uric acid molecules and one potassium atom in general positions. This implied that both independent molecules had to be half-deprotonated, this being realized through a disorder between two molecules related by an inversion center, only one being deprotonated randomly. The IUPAC name is proposed to be potassium 7,9-dihydro-3H-purine-2,6,8-trione 7,9-dihydro-1H-purine-2,6,8(3H)-trione. The final Rietveld [34] refinements were realized by using the FULLPROF software [35]. All non-H atoms may be refined freely, but this led to highly distorted molecules so that 102 geometric restraints were applied on the uric acid and urate molecules. These restraints were derived from the recent accurate single crystal structure determination of CaUr2·6H2O [16]. The two independent molecules in the title compound were treated during the structure refinements as having both the C5H3.5N4O3 formula, one hydrogen atom being half occupied. The half occupied H atom sites are close to the inversion centers relating two such molecules, only one having to be deprotonated forming the urate anion and the other being a uric acid molecule. Anisotropic line broadening was noticed and treated by using the in-built FULLPROF spherical harmonic options, concluding a negligible strain effect and nanometer size of the platelet-like coherently diffracting domains having the shortest mean size (290 Å) in the direction of the a axis, 650 Å along b and 500 Å along c. The Rietveld plot is shown in Fig. 1. The Rietveld refinement details are in Table 1, atom numbering in the two independent uric acid molecules is shown in Fig. 2, selected geometric parameters and the hydrogen bonds are shown in Tables 2 and 3.
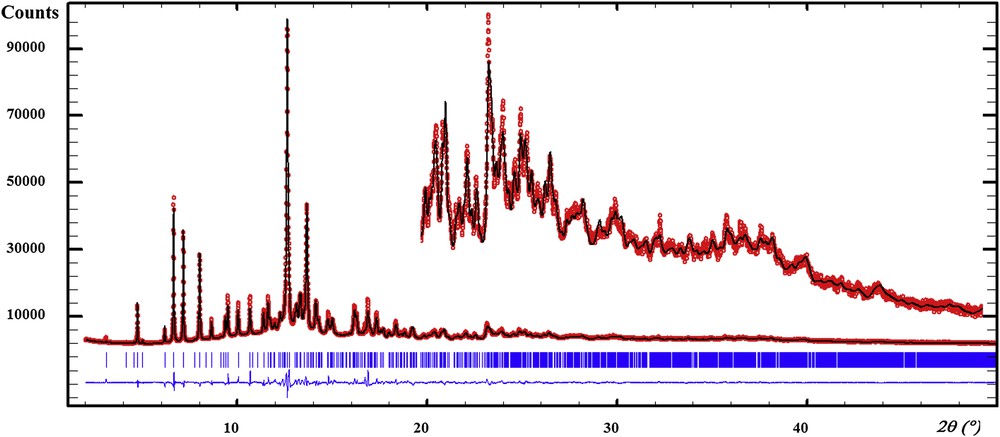
Refined synchrotron diffraction pattern for the potassium quadriurate. Red dots represent the observed data, the black line represents the calculated ones. Bragg ticks are the peak positions and the blue curve shows the difference between the observed and calculated patterns.
Synchrotron XRD experimental parameters and Rietveld refinement details of the potassium quadriurate.
| Wavelength (Å) | 0.72442 |
| Synchrotron | SOLEIL, France |
| Beamline | CRISTAL |
| Monochromator | Si(111) |
| Temperature (°C) | 23 |
| Scan range (°2θ) | 2–50 |
| Step size (°2θ) | 0.005 |
| Chemical formula | C10H7KN8O6 |
| Chemical formula moiety | K(C5H3N4O3)(C5H4N4O3) |
| Space group | P21/c |
| Z | 4 |
| Cell parameters (Å, °) | |
| a | 3.60504(9) |
| b | 19.9293(7) |
| c | 18.4841(6) |
| β | 100.503(2) |
| Volume (Å3) | 1305.75(7) |
| No. of contributing reflections | 2176 |
| No. of refined parameters (total) | 153 |
| No. of atomic coordinates | 99 |
| No. of restraints | 102 |
| No. of isotropic thermal parameters | 4 |
| No. of background parameters | 38 |
| Profile shape | pseudo-Voigt |
| η | 0.838(7) |
| U | 0.72(3) |
| V | −0.029(4) |
| W | 0.00116(16) |
| Conventional Rietveld reliability factors | (Rietveld, 1969) |
| RP (%) | 9.46 |
| RWP (%) | 10.9 |
| Rexp (%) | 3.55 |
| RB (%) | 3.28 |
| RF (%) | 1.93 |
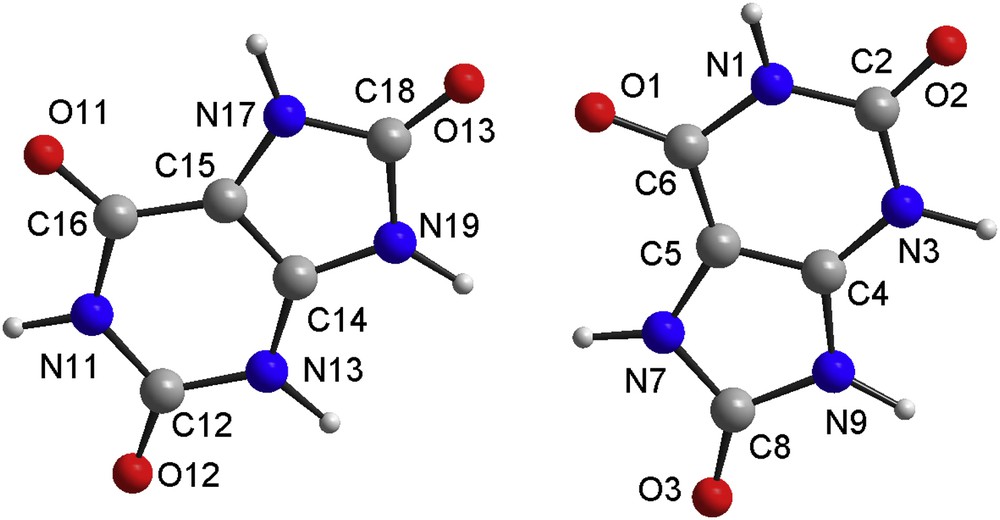
Atom numbering in the two independent uric acid molecules, being statistically half N3-deprotonated (or N13).
Selected geometric parameters (Å, °) for the potassium quadriurate. The DFT-optimized values are in italics.
| K–O3i | 2.855 (6) | 2.921 | N7–C5 | 1.396 (8) | 1.386 |
| K–O3ii | 2.828 (6) | 2.876 | N7–C8 | 1.354 (7) | 1.357 |
| K–O11iii | 2.709 (5) | 2.724 | N9–C4 | 1.393 (8) | 1.359 |
| K–O1iv | 2.817 (6) | 2.701 | N9–C8 | 1.391 (6) | 1.392 |
| K–O13iv | 2.766 (6) | 2.712 | N11–C12 | 1.378 (5) | 1.372 |
| K–O1v | 2.752 (6) | 2.684 | N11–C16 | 1.405 (7) | 1.394 |
| K–O13v | 2.627 (6) | 2.628 | N13–C12 | 1.355 (4) | 1.360 |
| O1–C6 | 1.249 (5) | 1.232 | N13–C14 | 1.349 (7) | 1.343 |
| O2–C2 | 1.273 (6) | 1.245 | N17–C15 | 1.405 (8) | 1.384 |
| O3–C8 | 1.245 (7) | 1.236 | N17–C18 | 1.352 (5) | 1.356 |
| O11–C16 | 1.254 (6) | 1.241 | N19–C14 | 1.380 (4) | 1.353 |
| O12–C12 | 1.286 (6) | 1.240 | N19–C18 | 1.391 (9) | 1.396 |
| O13–C18 | 1.228 (6) | 1.229 | C4–C5 | 1.386 (5) | 1.367 |
| N1–C2 | 1.371 (5) | 1.366 | C5–C6 | 1.396 (9) | 1.411 |
| N1–C6 | 1.386 (7) | 1.398 | C14–C15 | 1.380 (7) | 1.368 |
| N3–C2 | 1.368 (8) | 1.357 | C15–C16 | 1.394 (4) | 1.404 |
| N3–C4 | 1.346 (5) | 1.348 | |||
| C2–N1–C6 | 127.5 (5) | 127.61 | N3–C4–N9 | 125.8 (4) | 127.17 |
| C2–N3–C4 | 113.9 (4) | 118.87 | N3–C4–C5 | 125.7 (4) | 124.33 |
| C5–N7–C8 | 108.5 (4) | 109.23 | N9–C4–C5 | 105.9 (4) | 108.50 |
| C4–N9–C8 | 109.3 (4) | 108.58 | N7–C5–C4 | 108.6 (5) | 107.04 |
| C12–N11–C16 | 126.5 (3) | 127.12 | N7–C5–C6 | 129.8 (4) | 132.98 |
| C12–N13–C14 | 114.4 (4) | 119.01 | C4–C5–C6 | 120.1 (4) | 119.94 |
| C15–N17–C18 | 108.2 (5) | 109.43 | O1–C6–N1 | 121.2 (6) | 120.22 |
| C14–N19–C18 | 108.9 (4) | 108.41 | O1–C6–C5 | 127.0 (4) | 127.49 |
| C6–N1–H1 | 114.9 (12) | 115.00 | N1–C6–C5 | 111.4 (3) | 112.29 |
| C2–N1–H1 | 117.0 (8) | 117.00 | O3–C8–N7 | 125.1 (4) | 127.04 |
| C4–N3–H3 | 121.6 (7) | 122.00 | O3–C8–N9 | 126.0 (5) | 126.32 |
| C2–N3–H3 | 115.2 (12) | 119.00 | N7–C8–N9 | 107.5 (5) | 106.64 |
| C5–N7–H7 | 124.0 (12) | 128.00 | O12–C12–N13 | 118.8 (4) | 121.53 |
| C8–N7–H7 | 125.3 (16) | 123.00 | N11–C12–N13 | 118.3 (3) | 116.82 |
| C8–N9–H9 | 125.5 (12) | 128.00 | O12–C12–N11 | 117.1 (3) | 121.64 |
| C4–N9–H9 | 122.8 (17) | 124.00 | N13–C14–N19 | 126.6 (5) | 127.06 |
| C12–N11–H11 | 116.9 (13) | 116.00 | N13–C14–C15 | 125.8 (3) | 124.08 |
| C16–N11–H11 | 112.7 (11) | 116.00 | N19–C14–C15 | 106.8 (5) | 108.86 |
| C14–N13–H13 | 119.1 (10) | 118.00 | N17–C15–C14 | 107.9 (3) | 107.77 |
| C12–N13–H13 | 116.8 (7) | 122.00 | N17–C15–C16 | 130.4 (6) | 133.02 |
| C15–N17–H17 | 123.9 (13) | 127.00 | C14–C15–C16 | 120.2 (5) | 120.20 |
| C18–N17–H17 | 127.7 (15) | 123.00 | O11–C16–C15 | 127.6 (5) | 126.77 |
| C14–N19–H19 | 125 (2) | 125.00 | N11–C16–C15 | 109.9 (5) | 112.68 |
| C18–N19–H19 | 126 (2) | 127.00 | O11–C16–N11 | 120.0 (4) | 120.55 |
| O2–C2–N1 | 119.5 (5) | 121.5 5 | O13–C18–N19 | 126.1 (4) | 126.49 |
| O2–C2–N3 | 120.9 (4) | 121.53 | N17–C18–N19 | 107.8 (4) | 106.53 |
| N1–C2–N3 | 118.6 (5) | 116.91 | O13–C18–N17 | 125.9 (7) | 126.99 |
Hydrogen geometry (Å,°) for the potassium quadriurate. The DFT-optimized values are in the second line (italics).
| D–H⋯A | D–H | H⋯A | D⋯A | D–H⋯A |
| N1–H1⋯O11i | 0.90 (2) 1.030 | 2.18 (4) 1.890 | 2.985 (8) 2.894 | 150 (5) 164.0 |
| N3–H3⋯N3ii | 0.893 (10) 1.100 | 1.626 (9) 1.610 | 2.500 (4) 2.708 | 165 (2) 179.0 |
| N7–H7⋯O12iii | 0.882 (19) 1.030 | 2.040 (17) 1.800 | 2.880 (5) 2.923 | 159 (2) 172.0 |
| N9–H9⋯O2ii | 0.804 (13) 1.030 | 2.187 (12) 2.020 | 2.890 (5) 2.992 | 146.3 (12) 157.0 |
| N11–H11⋯O3iii | 0.90 (2) 1.030 | 2.18 (2) 1.920 | 3.006 (5) 2.949 | 152 (2) 173.0 |
| N13–H13⋯N13iii | 0.900 (15) 1.100 | 1.893 (17) 1.550 | 2.517 (5) 2.645 | 124.7 (14) 174.0 |
| N17–H17⋯O2iv | 0.87 (4) 1.030 | 1.92 (4) 1.730 | 2.766 (8) 2.757 | 164 (3) 177.0 |
| N19–H19⋯O12iii | 0.80 (2) 1.030 | 2.15 (3) 1.880 | 2.908 (7) 2.888 | 160 (3) 164.0 |
4 Solid state DFT structure optimization
All the solid state calculations were performed using the VASP code [36–38]. To optimize the structures, the hybrid PBE0 functional [39] was used. Plane waves formed a basis set and the calculations were performed using the projector-augmented wave method [40,41] and atomic pseudo-potentials [42]. The energy cutoff controlling the accuracy of structure optimization calculations was in both cases set to 700 eV and the Brillouin zones were sampled in four k-points. The positions of all 128 atoms (C40H28K4N32O24) in the computational P1 space group description of the structure were fully relaxed while keeping the lattice parameters of the respective cells fixed at the values obtained from diffraction data. Half occupied hydrogen atoms used in Rietveld refinement were replaced by localized hydrogen atoms placed arbitrarily on respective N-atoms. Geometrical analysis of the optimized structures done by means of the PLATON program [43] showed that the originally proposed space group was correct. It has been previously proved that the DFT-optimized structure provides intermolecular distances certainly more accurate than those resulting from the Rietveld refinement which restrained the intramolecular interatomic distances to a quasi-rigid model [see, e.g., 44,45]
5 Discussion
The organic moieties exhibit C–C, C–N and C–O bond distances and angles obtained by minimization of the total energy (Table 2), which match well those in uric acid obtained from single crystal X-ray data [11]. Both molecules are essentially planar with the maximum deviation from the planes fit to all non-hydrogen atoms of 0.05 Å and 0.1 Å, respectively. The projection of the structure along the short a axis (Fig. 3) shows that every uric acid molecule is connected to a urate anion through hydrogen bonds.
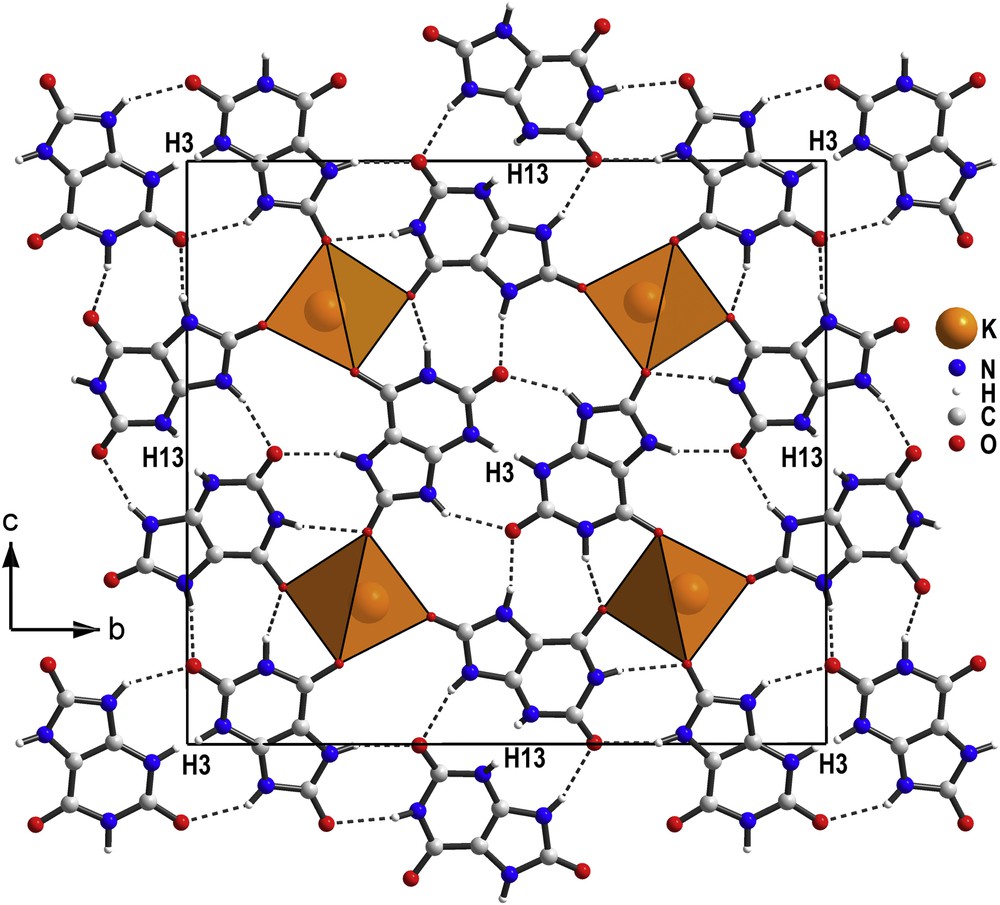
Projection along the short a axis of the potassium quadriurate structure showing the KO7 monocapped trigonal prisms and the pairs of urate and uric acid molecules. The hydrogen atoms H3 and H13, close to inversion centers, are half occupied, then choosing one position decides for which molecule is deprotonated or not.
Potassium quadriurate is thus a co-crystal since it is made of 1:1 M combination of uric acid and potassium urate. Nevertheless, the uric acid molecule and the urate anion are probably randomly distributed. This implies that the centers of inversion are pseudo ones unless H3 and H13 are placed exactly on them. Apart from the dominating electrostatic cation–anion interactions, the pairs uric acid-urate also form N–H…N and N–H…O hydrogen bonds (Table 3). The N…N distances in these strong hydrogen bonds obtained by solid state DFT structure optimization compare 2.633(7) Å in 9-ethylguanine hemihydrochloride [46], 2.692(4) Å in a supramolecular assembly of phthalimide and 2-guanidinobenzimidazole [47] or to 2.724(4) Å in d,l-histidyl-L, d-histidine pentahydrate [48]. When considering the large D–H…A angles, the N–H…O bonds in the structure belong to stronger hydrogen bonds. The D…A distances well-correspond to the maximum on the histogram (2.892(3) Å) constructed by using 1357 intermolecular N–H…OC bonds [49]. The oxygen atoms O2 and O12, which are not involved in the potassium coordination sphere, are double acceptors of hydrogen bonds (Fig. 4).
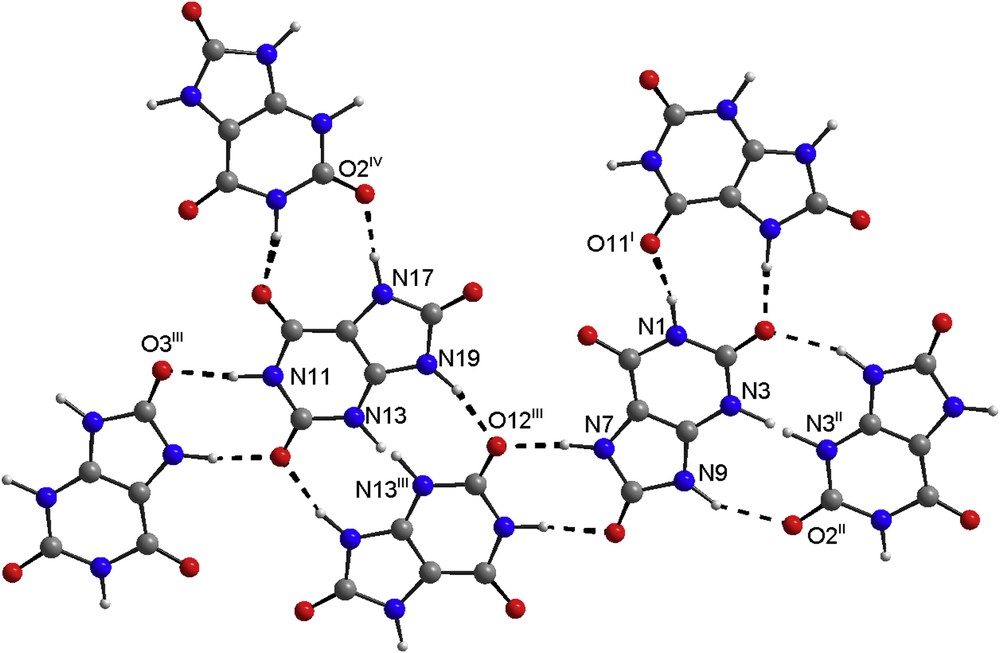
Hydrogen bonds are shown by dashed lines, symmetry codes are defined in Table 3. Note that H3 and H13 atoms attached to N3/N13 are shown in both possible positions. (In the color version nitrogen atoms are in blue, oxygen in red and carbon in gray).
In the CaUr2·6H2O structure [16], only one of the two urate molecules is directly coordinated to Ca2+ in a bidentate fashion through N3 and O2, six water molecules completing the CaO7N polyhedron. In the potassium quadriurate the K+ ion is seven-coordinated by oxygen atoms from seven different molecules of either urate or uric acid. Only two of the oxygen atoms of the uric acid molecules and two of the urate anions (O1, O3 and O11, O13 with K–O distances < 3 Å) are involved in the potassium coordination forming KO7 monocapped trigonal prisms sharing their triangular basis, building infinite chains extending along the short a axis. This kind of cell with a very short parameter is characteristic of compounds with parallel stacking of molecules. Interestingly, theoretical study of interactions of urate with K+ in vaccuo predicts a minimum of energy when the cation interacts with oxygen and nitrogen of the six-membered ring [50]. In the structure of the title compound both these atoms are involved in hydrogen bonds and hence sterically blocked.
Analogous urate molecule stacking is known in sodium urate monohydrate (short axis = 3.567 (1) Å) with infinite chains of NaO6 octahedra sharing edges along that short axis [14]. Such a short axis is known too in the case of ammonium urate (frequent in urolithiasis in Dalmatians) for which a monoclinic cell with b = 3.528(1) Å was proposed [51] as well as a triclinic one with a = 3.650(3) Å [15] from powder data indexing. Such a close stacking (3.664(1) Å) of molecules is also the case for purine [52]. Anyway, the sodium urate crystallizes in needles elongated along this short axis, contrarily to the quadriurate of potassium for which this direction is apparently the most difficult for crystal growth (platelets).
6 Conclusion
To our knowledge, this is the first observation of such a combination by hydrogen bonding of one uric acid molecule with one N3-deprotonated molecule. We have to recognize the strong expertize of the XIXth century scientists who proposed the quadriurate's existence well before any X-ray crystallography possibility of characterization. Sir William Roberts in 1892 [23] asked “what is the chemical nature of this substance, what are its physiological analogies and pathological relations? What is the constitution of this substance? Is it merely a mechanical mixture of biurate with varying quantities of free uric acid or a new and third order of uric acid salts, differing essentially from the two regular orders previously recognized?” In spite of Kohler [24] declaring that no such body as quadriurate exists, we have answered here to the first part of the question and established the reality of the potassium quadriurate as belonging to a third order of uric acid salts, thanks to powder diffraction modern methodologies allowing crystal structure determination. The possible existence of sodium or ammonium quadriurates is now questioned but highly probable (their mixture with potassium quadriurate being the main constituent of bird and snake excretions). Finally, the naming is confusing since the still crystallographically uncharacterized KC5H3N4O3 is generally called potassium urate, then the title compound would logically be a biurate in the modern nomenclature, not a quadriurate. Concerning physiological implications, the current views about the excretion of uric acid, which is very poorly soluble, are that the urates, which are more soluble, pass into the excretory tubules and that the urine is later acidified. The quadriurate of potassium, highly soluble in urines, provides a solution for the elimination of uric acid which would crystallize only after the potassium quadriurate degradation into potassium urate and uric acid in water. An explanation for the efficiency of potassium citrate on urolithiasis may come from the way a urate anion can link by three hydrogen bonds to a uric acid molecule and form a new bigger anion compensated by only one K+ cation (or Na+, NH4+, etc). This fact may well lead to the formulation of a new hypothesis about urate transport, a subject having attracted considerable interest in the past 150 years [53].
Acknowledgments
The authors thank Pierre Gras and Kémie Ley-Ngardigal for technical assistance with the recording of the synchrotron pattern. Calculations were performed in the Computing Centre of the Slovak Academy of Sciences using supercomputing infrastructure acquired within the project ITMS 26230120002 and 26210120002 (Slovak Infrastructure for High-Performance Computing) supported by the Research and Development Operational Programme funded by the ERDF. Support of VEGA 2/0131/12 is appreciated.
Appendix A Supplementary data
The following is the supplementary data related to this article:
Full crystallographic data for the structural analysis have been deposited with the Crystallographic Open Database, COD No. 3000040 and with the Cambridge Crystallographic Data Center, CCDC No. 1042936. These data can be obtained free of charge via www.crystallography.net or via www.ccdc.cam.ac.uk or from Cambridge Crystallography Data Center, 12, Union Road, Cambridge, CB2, 1EZ, UK (fax: +44 1223 336 033 or email: deposit@ccdc.cam.ac.uk).


