1 Introduction
Thiosemicarbazone molecules with nitrogen and sulfur donor atoms are important ligands in coordination chemistry [1,2]. Their complexes with a transition metal have interesting biological and chemical properties [1,2], variable bonding features, and considerable structural diversity [3–11]. The biological properties of thiosemicarbazone complexes are often related to the coordination pathway towards the transition metal ion [2–5]. Thiosemicarbazones bind to the transition metal center in a neutral or anionic form [1–11]. Variable bonding and structural diversity of metal complexes with thiosemicarbazone derivatives were reviewed in 2009 by Lobana and his co-workers [12]. Copper(I) forms both charged and neutral complexes with thiosemicarbazone ligands [13–23]. Moreover, it was found that the substituents at the carbon atom of the imine group (CN) affect the bonding to the copper(I) halides, determining the formation of mono-, di- and poly-nuclear complexes. Often, these complexes crystallize in the presence of PPh3 as a co-ligand.
Following our previous work on thiosemicarbazone complexes [24–26], here we report a simple ultrasonic-bath-assisted preparation of (nano)particles of a mononuclear copper(I) complex [Cu(Brcatsc)(PPh3)2Cl]·CH3CN (1) (Scheme 1), which is synthesized by the reaction of CuCl, Brcatsc and PPh3 (molar ration 1:1:2).
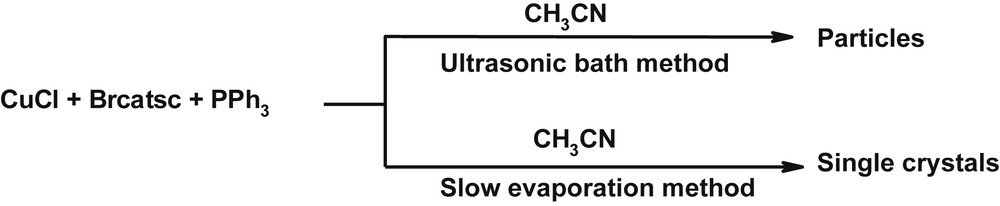
Materials produced and synthetic methods.
2 Experimental
2.1 Reagents and physical measurements
All reagents and solvents for synthesis and analysis were obtained from Merck and used as received. The thiosemicarbazone ligand Brcatsc was prepared in high yield following the literature [27]. The FT-IR spectrum was recorded in the range 400–4000 cm−1 on a PerkinElmer FT-IR spectrophotometer using KBr pellets. Elemental analyses were performed on a Heraeus CHNO-Rapid analyzer and the results agreed with the calculated values. 1H, 13C and 31P NMR spectra were recorded on a BRUKER DRX-400 AVANCE spectrometer at 300 MHz for the Schiff base ligand. All chemical shifts are reported in δ units downfield from tetramethylsilane (TMS). The thermogravimetric analysis (TG) was performed in the temperature range 30–750 °C on a PerkinElmer TG/DTA lab system 1 (technology by SII). The samples were held in a nitrogen atmosphere and a heating rate of 20 °C/min was set. X-ray powder diffraction was performed on a Philips X'pert diffractometer using Cu Kα radiation. The SEM images were obtained using a Philips XL 30 scanning electron microscope. The samples were made conductive by a thin gold coating. UV–vis spectra were recorded on a PerkinElmer Lambda 25 spectrometer.
2.2 Antibacterial activity
The antibacterial performance of Brcatsc and its copper(I) complex 1 was investigated against two gram-positive (Staphylococcus aureus ATCC-25923 and Enterococcus faecalis ATCC-29212) and two gram-negative (Escherichia coli ATCC 25922 and Pseudomonas aeruginosa ATCC-27853) bacteria. The tests were performed using a methodology described in the guidelines of the Comité de l'Antibiogramme de la Société Francaise de Microbiologie (CA-SFM, www.sfm.asso.fr) [28]. A solution of Brcatsc and 1 was prepared at 20 mg/mL in DMSO under sterile conditions. After dilution, 10 Petri plates were prepared with concentrations from 10 to 100 μg/mL (increasing by 10 μg/mL). The different Petri plates were inoculated by ≈2 × 104 bacteria suspended in sterile distilled water. After 24 h incubation at 37 °C, the minimum inhibitory concentration (MICs, μg/mL) was determined.
2.3 X-ray structure determination
A single crystal of dimensions 0.11 × 0.08 × 0.07 mm3 was chosen for the X-ray diffraction study. Crystallographic measurements were performed at 120 K with a four-circle CCD diffractometer Gemini (Oxford diffraction, Ltd.), equipped with mirror-collimated Cu Kα radiation (λ = 1.54184 Å). Although the quality of the single crystals was not the best (as indicated, e.g., by the Rint factor in Table 1), the crystal structure could be easily solved by the charge flipping program SUPERFLIP [29]. The refinement was carried out by the Jana2006 program package [30] employing a full-matrix least-squares technique on F2. The molecular structure plots were prepared by Diamond 4.0 [31]. The hydrogen atoms were mostly discernible in difference Fourier maps and could be refined to a reasonable geometry. According to common practice, the hydrogen atoms attached to carbons were kept in ideal positions during the refinement with a CH distance of 0.96 Å and their isotropic atomic displacement parameters were set to 1.2Ueq of their parent atoms. Crystallographic data, details of the data collection, structure solution, and refinements are listed in Table 1.
Crystallographic data and structural refinement details of 1.
| Empirical Formula | C48H41Br1Cl1Cu1N4P2S1 |
| Formula weight | 946.8 |
| Crystal system, space group | Triclinic, |
| a, Ǻ | 10.5899(4) |
| b, Ǻ | 13.4301(6) |
| c, Ǻ | 17.0554(8) |
| α, deg | 68.874(4) |
| β, deg | 80.139(3) |
| γ, deg | 88.483(3) |
| V, Ǻ3 | 2227.58(18) |
| Z | 4 |
| μ, mm−1 | 5.52 |
| Measured/independent reflections | 12,950/7586 |
| Rint | 0.084 |
| Reflections with I > 3σ(I) | 7822 |
| Parameters | 1045 |
| S | 1.52 |
| R(F2 > 3σ(F2)) | 0.043 |
| wR(F2) | 0.053 |
This complex has disordered acetonitrile molecules. Two positions of this linear molecule, with refined occupancy of 0.775(12) and 0.225(12), are oriented in such a way that they almost coincide on one side but are considerably deviated on the opposite side. The decision whether the overlapped part combines two close nitrogen atoms or one nitrogen and one CH3 group was further complicated by a smeared difference in the electron density in this area without clear indication of the hydrogen atoms. Refinement of two structure models indicated that the two disordered acetonitrile molecules have probably opposite orientations, combining N and CH3, because this model yielded a geometry closer to linear and better interatomic distances.
2.4 Preparation of 1
2.4.1 Slow evaporation
Solid PPh3 (0.052 g, 0.2 mmol) was added to CuCl (0.009 g, 0.1 mmol) suspended in acetonitrile (10 mL), and the mixture was stirred for 0.5 h. To this mixture, the Brcatsc ligand in the solid state was added (0.028, 0.1 mmol), and the content was stirred until a clear solution was obtained. The solution was allowed to evaporate slowly at room temperature for several days. The white polygonal crystals were filtered, washed twice with acetonitrile, and dried at room temperature (0.65 g, 75.6%). Anal. calcd. for C48H41Br1Cl1Cu1N4P2S1: C, 60.89; H, 4.33; N, 5.92; S, 3.38%. Found: C, 60.38; H, 4.8; N, 5.77; S, 4.08%. FT-IR data (KBr, cm−1): υ (NH) 3150, υ (NH2 group), 3264–3427, υ (CH aromatic) 3049, υ (CH imine) 2802, υ (CN imine) 1591, υ (CC aromatic) 1537, { υ (CN) + υ (CC) + υ (PCph)} 1028, 1097, 1175, υ (CS) 854, υ (CuS) 510. 1H NMR (DMSO-d6, δ ppm): 11.9 (s, 1H, NH), 8.6 (s, 1H, CbH), 7.9 (d, 1H, ChH), 7.6 (d, 2H, NH2), 7.2–7.4 (m, 31H, PPh3 + CdH), 7.0–7.1 (d, 2H, Cf,jH), 6.8–6.9 (dd, 2H, Cg,iH). 13C NMR (DMSO-d6, δ ppm): 176.1 (CaS), 146.6 (CbN), 125.0–140.1 (Cc–j + PPh3). 31P NMR (DMSO-d6, δ, ppm): −5.5.
2.4.2 Ultrasonic bath
In order to prepare particles of 1, 10 mL of CuCl (0.1 mmol) in acetonitrile was placed in an ultrasonic bath, with a power output of 40 KHz. Into this solution, a 10 mL solution of Brcatsc (0.1 mmol) and PPh3 (0.2 mmol) was added dropwise. The obtained precipitate was filtered off, washed with acetonitrile and then dried in air (0.78 g, 90.7%). FT-IR data (KBr, cm−1): ν(NH2 group) 3418, 3266, ν(NH) 3137, ν(CH aromatic) 3050, ν(CN imine) 1592, ν(CC) 1537–1479, ν(CN) 1096, ν(CS) 932, 853.
3 Results and discussion
3.1 Synthesis and spectroscopic characterization
The reaction of Brcatsc with an acetonitrile solution of copper(I) chloride (CuCl) (1:1 molar ratio), followed by the addition of triphenyl phosphine (PPh3), yielded bright yellow crystals of the mononuclear copper(I) thiosemicarbazone complex of empirical formula [Cu(Brcatsc)(PPh3)2Cl]·CH3CN (1) (Fig. 1). The elemental analysis revealed that the ratio of Brcatsc, PPh3 and CuCl was 1:2:1. To our knowledge this is the first copper(I) thiosemicarbazone complex containing bromocinnamaldehyde.
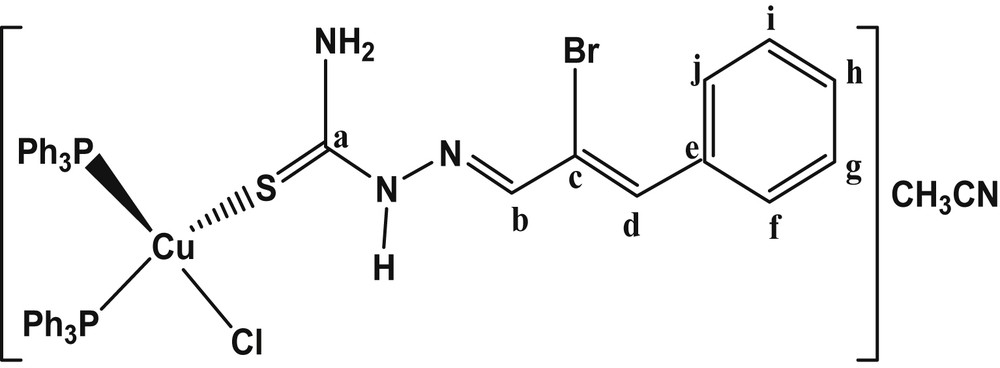
The chemical structure of 1.
The FT-IR spectrum of the complex (Fig. 2) is similar to the one of the free ligand [27] along with some distinct changes in the position of the bands. The presence of three ν(NH) bands at 3150 cm−1 (due to NNHC) and 3264–3427 cm−1 (due to NH2) indicates the coordination of a neutral ligand in the complex. In addition, the characteristic bands of ν(CS) and ν(CN) vibrational modes as seen at 854 and 1591 cm−1, respectively, indicate the presence of the ligand. The shifting of the ν(CS) band to a higher energy in the complex compared to the free ligand (859 cm−1) confirms that the Brcatsc ligand is coordinated to copper(I) by the S-donor atom. Further, ν(CH aromatic) and {ν(PCPh) + ν(CC) + ν(CN)} bands are present at 3049 cm−1 and in the range 1028–1175 cm−1, respectively [16,18,25]. As shown in Fig. 2, the FT-IR spectra of the complex 1 prepared by the two different methods are similar.
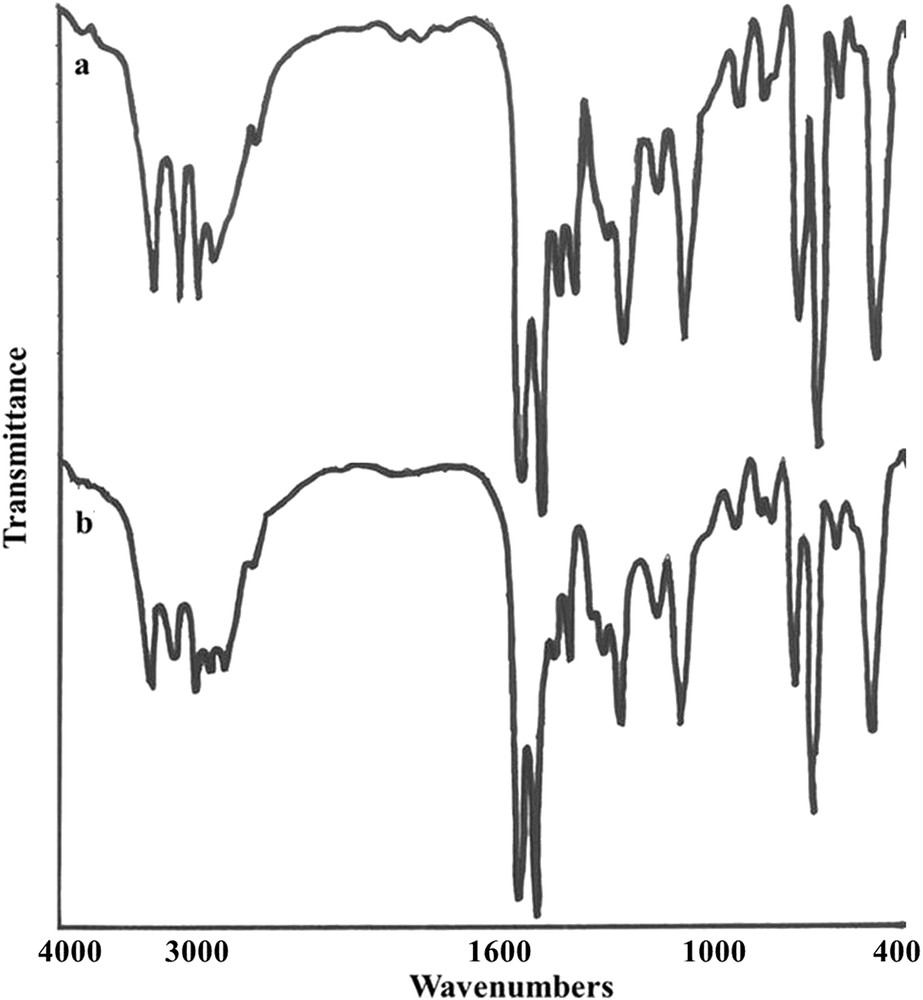
FT-IR spectrum of 1 prepared by two different routes: a) slow evaporation and b) ultrasonic bath.
1H, 13C and 31P NMR spectra of 1 were recorded using DMSO-d6 as the solvent and are shown in Fig. 3. The 1H NMR spectrum of 1 (Fig. 3a) exhibited the NNHCS singlet signal at about 11.5 ppm, indicating coordination of the ligand to the copper(I) ion in its neutral form, probably via the S-donor atom [25,26]. A singlet signal ≈ 8.5 ppm also reveals the presence of the iminic hydrogen (CHN). The aromatic protons of Brcatsc in 1 appear as three signals at 7.0–7.1 (d, 2H, Cf,jH, J = 16 Hz), 6.8–6.9 (dd, 2H, Cg,iH, J = 16 Hz) and 7.9 ppm (d, 1H, ChH, J = 8 Hz). The ethylenic proton (CdH) and aromatic protons of PPh3 are present in the range 7.26–7.41 ppm (m, 31H, PPh3 + CdH). The amine protons (NH2) of Brcatsc appeared as doublet signals at about 7.90 ppm. The aromatic protons of PPh3 appear between 7.26 and 7.41 ppm [25,26].
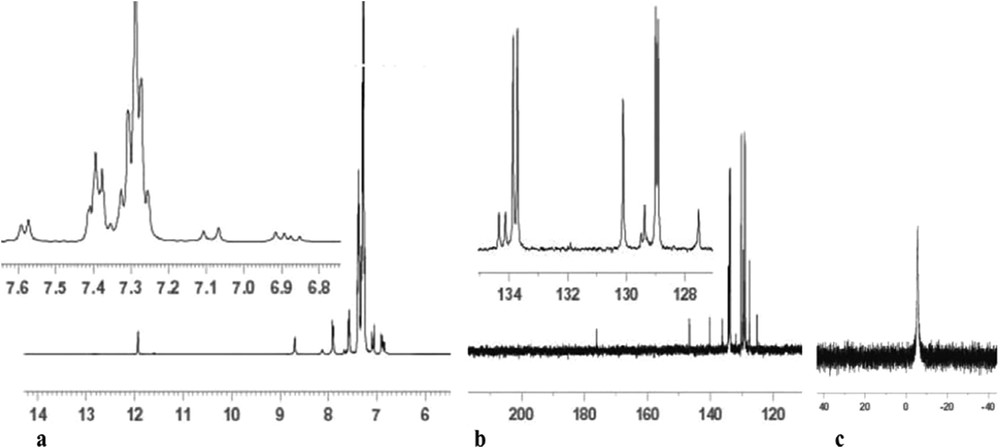
NMR spectra of 1: a) 1H, b) 13C, and c) 31P.
The 13C NMR spectrum (Fig. 3b) also shows signals for the carbon atoms of CaS and CbN groups at 175 and 145 ppm, respectively. Other carbons (aromatic and ethylenic) can be seen as signals in the interval 120–145 ppm. The 31P NMR spectrum of 1 (Fig. 3c) shows only a singlet peak at 5.55 ppm for the two P atoms, which confirms their equivalent chemical environment. Moreover, it points that the geometry around the copper(I) ion is tetrahedral and not square-planar, which is in agreement with the bonding angles involving the copper(I) ion ranging from 102.48 to 125.11°.
3.2 Crystal structure of 1
Complex 1 crystallizes in the triclinic crystal system (space group ). The triclinic unit cell contains two molecules of the complex. The molecular structure of 1 is depicted in Fig. 4. Selected bond distances and angles are presented in Table 2. In 1, the copper(I) ion is bonded to one chlorine atom, one sulfur atom (of the Brcatsc ligand), and two phosphorous atoms from two PPh3 groups, forming a distorted tetrahedral geometry around the Cu(I) center. The bond distances Cu1S1 (2.3650(16) Å), Cu1Cl1 (2.3911(15) Å), Cu1P1 (2.2738(18) Å) and Cu1P2 (2.2586(18) Å) are similar to those reported for other mononuclear copper(I) thiosemicarbazone complexes [15–17,24]. Bond distances and different bond angles are given in Table 2 and confirm the distorted tetrahedral geometry around the copper(I) center in 1 [15–17,24].
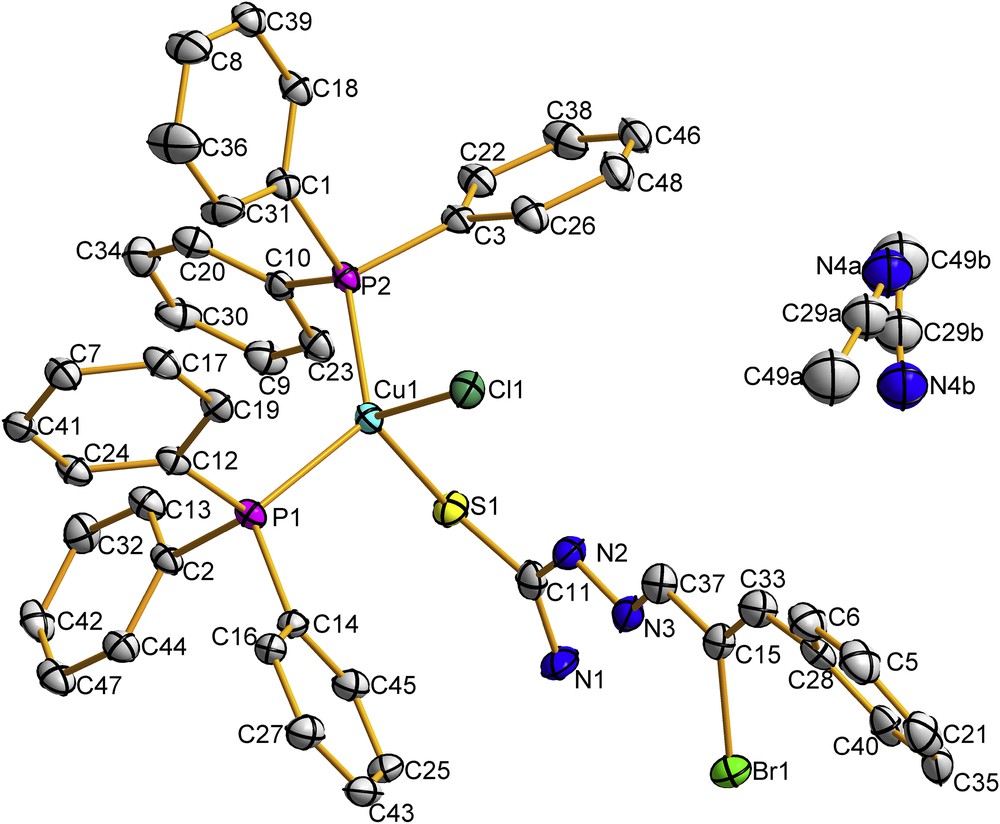
Molecular structure of 1 with atomic displacement ellipsoids shown at a 40% probability level. H atoms are omitted for clarity. The disorder of the acetonitrile is explained in Section 2.4.
Selected bond distances (Å) and angles (°) of 1.
| Bond lengths in Å | Bond angles in degrees | ||
| Cu1Cl1 | 2.3911(15) | Cl1Cu1P1 | 103.70(5) |
| Cu1S1 | 2.3650(16) | Cl1Cu1P2 | 108.02(5) |
| Cl1N2 | 3.129(6) | P1Cu1P2 | 125.11(7) |
| S1C11 | 1.689(7) | Cl1Cu1S1 | 111.72(6) |
| N1C11 | 1.324(9) | P1Cu1S1 | 105.75(5) |
| N2C11 | 1.364(7) | S1Cu1P2 | 102.48(6) |
| Cu1P1 | 2.2738(13) | Cl1N2C11 | 118.2(4) |
| Cu1P2 | 2.2586(18) | N2N3C37 | 115.4(6) |
| N2N3 | 1.362(9) | Cl1N2N3 | 119.1(3) |
| N3C37 | 1.279(7) | N3N2C11 | 119.3(5) |
| Cu1Cl1N2 | 74.39(10) |
As mentioned above, the bond angles around Cu vary from 102.48 to 125.11°, indicating a distorted tetrahedral geometry. The maximum deviation from 109.5° was found for the P1Cu1P2 angle (125.11°) and can be explained by the presence of six aromatic rings in the two PPh3 [15–17]. The sulfur atom S1 and the hydrazinic nitrogen N3 are in the E position with respect to the C11N2 bond. The stability of this configuration in the thiosemicarbazone ligand is due to the presence of an intra-molecular hydrogen bond (N2H1⋅⋅⋅Cl1). The coordination of the Brcatsc ligand to copper(I) via the sulfur atom causes a decrease of the CS bond distance in the complex (1.689(7) Å) compared to the free ligand (1.702(5) Å) [26].
3.3 TGA of 1
To examine the thermal stability of 1, thermogravimetry (TG) was carried out between 30 and 750 °C under a nitrogen flow (Fig. 5). The compound 1 was found to be stable up to about 250 °C. Decomposition of 1 occurs between 250 and 700 °C with a total mass loss of 85%.
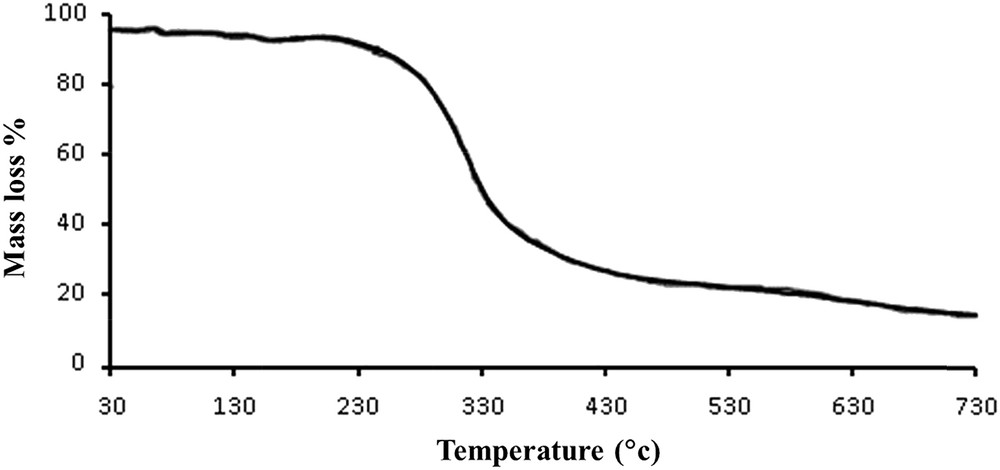
Thermogram of 1.
3.4 XRD and SEM of 1
Fig. 6 shows the XRD pattern of 1 prepared in an ultrasonic bath. The relatively sharp peaks indicates that the particles are of micrometric size and that the Scherrer formula can be used for calculation of the crystallites size forming the particles. Fig. 7 shows the scanning electron microscopy of 1 at two different scales. The SEM images show uniform morphology of the aggregated particles.
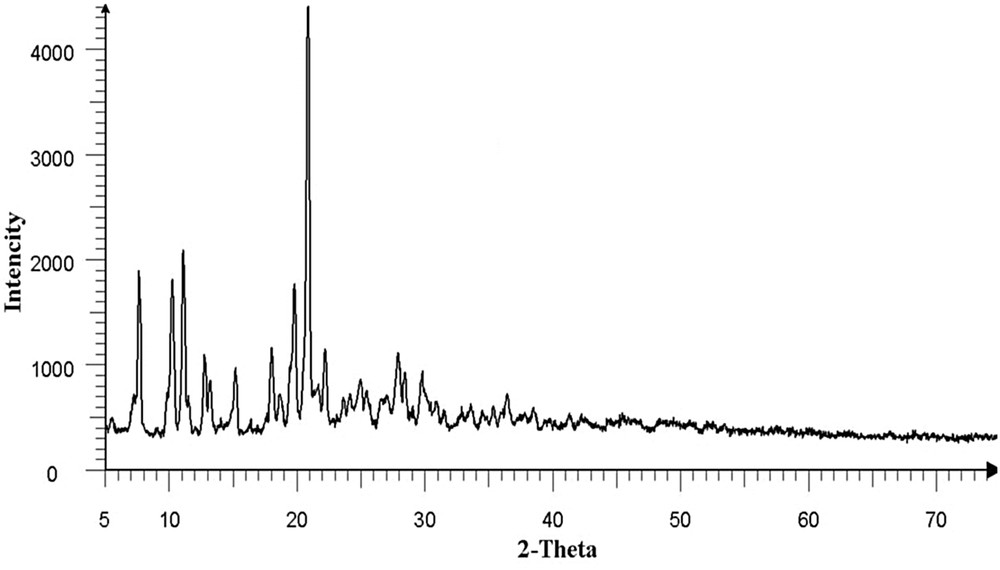
XRD pattern of 1 prepared in an ultrasonic bath.
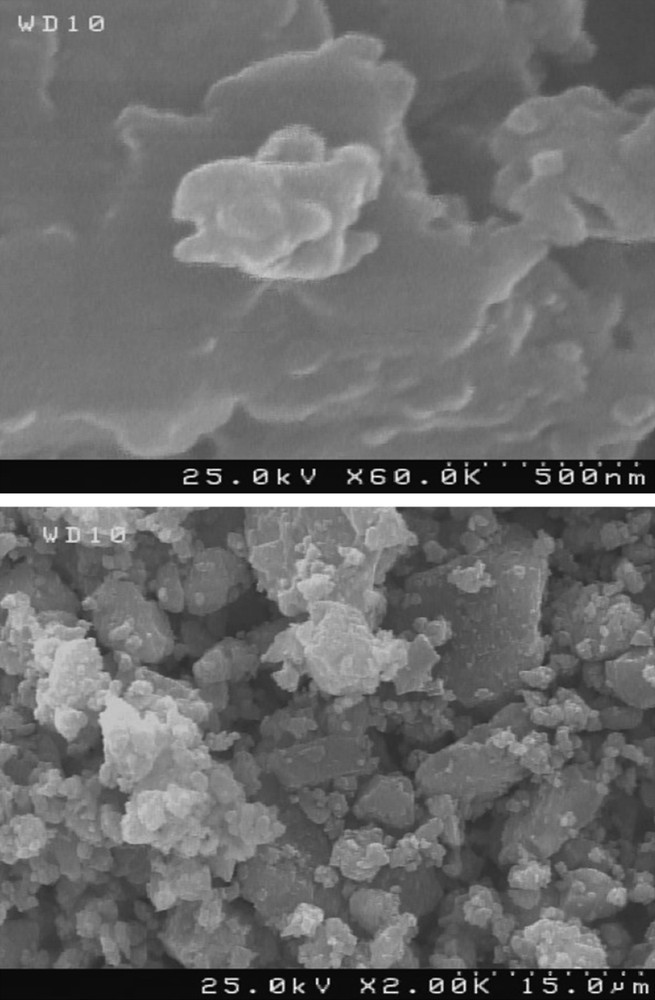
SEM images of 1 prepared in an ultrasonic bath.
3.5 Antibacterial activity
The antibacterial activity of Brcatsc and its copper(I) complex prepared by the two methods discussed above was studied against gram-positive (E. faecalis and S. aureus) and gram-negative (E. coli and P. aeruginosa) bacteria. The results show that the Brcatsc has no antibacterial activity against any of the tested bacteria even in concentrations as high as 500 μg/mL, while the complex is active against the gram-positive bacterial strains. The tested Petri plates are shown in Fig. 8. The MIC value (Table 3) of the complex against both E. faecalis and S. aureus is 70 μg/mL. The increased antibacterial activity of 1 compared to that of Brcatsc is due to the difference in the structures, which influences the attack on the cell walls of the bacteria [32,33]. In addition, the antibacterial activity of the complex prepared by two different methods was found to be similar.
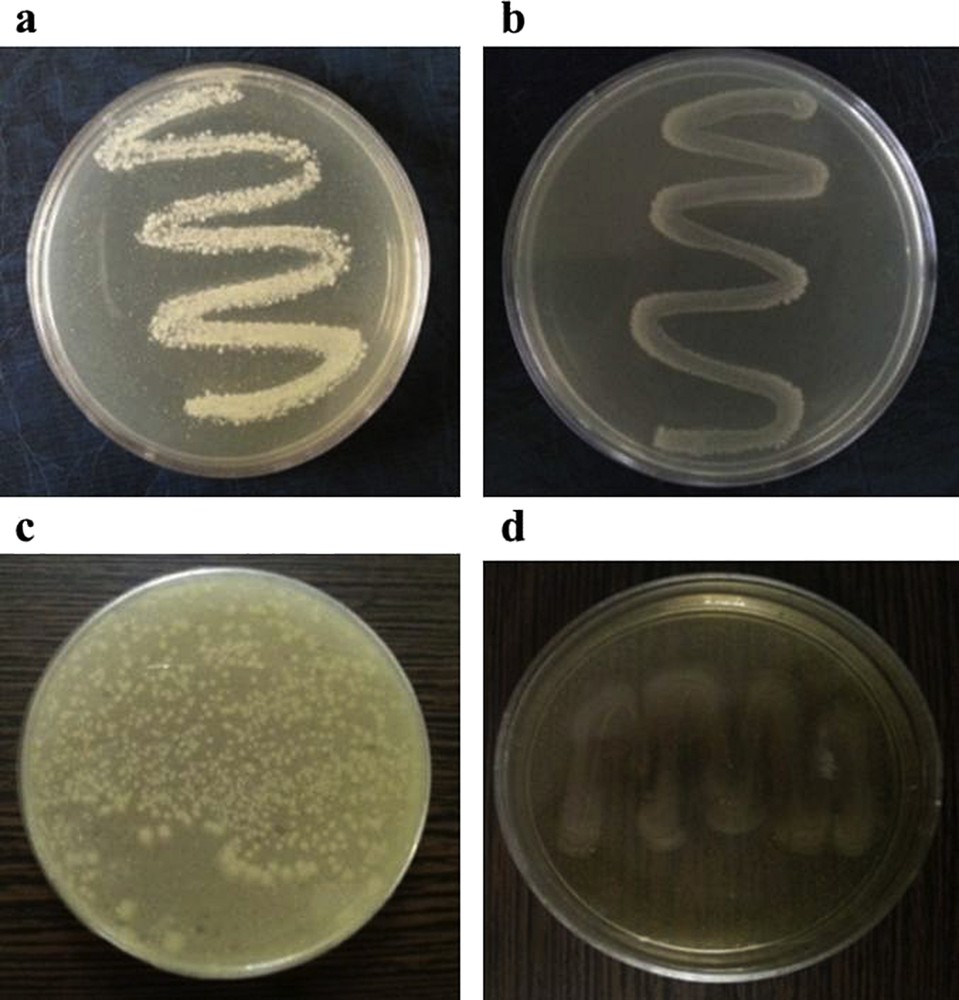
The antibacterial screening of 1 with 60 μg/mL concentration against a) S. aureus, b) E. faecalis, c) E. coli and d) P. aeruginosa.
The MIC value (μg/mL) of the ligand and the complex.
| MIC of the complex | MIC of the Brcatsc ligand | |
| E. faecalis (ATCC29212) | 70 | >500 |
| S. aureus (ATCC25923) | 70 | >500 |
| E. coli (ATCC25922) | >500 | >500 |
| P. aeruginosa (ATCC2753) | >500 | >500 |
4 Conclusion
A new copper(I) complex containing thiosemicarbazone was synthesized and characterized. Single-crystal structure determination showed that the copper(I) ion is coordinated with one S atom (of Brcatsc), one Cl and two P atoms in a distorted tetrahedral geometry. The spectroscopy data confirmed that the Brcatsc and PPh3 coordinate the copper(I) ion. An antibacterial screening indicated that the copper(I) complex exhibits considerably higher antibacterial activity than the free ligand towards gram-positive bacteria.
Acknowledgments
We are grateful to the Payame Noor University and Golestan University for financial support of this work. The crystallography was supported by the project 15-12653S of the Czech Science Foundation using instruments of the ASTRA lab established within the Operation program Prague Competitiveness–project CZ.2.16/3.1.00/24510.
Supplementary data
Crystallographic data (excluding structure factors) for the structure reported in this paper have been deposited with the Cambridge Crystallographic Center, CCDC No. 1042028. Copies of the data can be obtained free of charge on deposit@ccdc.cam.ac.uk or http:www.ccdc.cam.ac.uk.


