Version française abrégée
La faisabilité d'une enclave génétique propageant dans une cellule bactérienne certains messages héréditaires portés par une catégorie de polynucléotides distincte du DNA et du RNA est abordée dans cette note. L'analogue HNA choisi pour l'étude comporte un motif hexacyclique 1,5-anhydro-hexitol (menant aux hexitol-nucléotides) en remplacement du motif pentacyclique ribose ou 2-désoxyribose. Parmi les différents analogues de polynucléotides qui ont été élaborés en tant qu'agents antisens ou en tant que précurseurs prébiotiques des acides nucléiques, le HNA offre des caractéristiques fort propices à un lancement in vivo : il présente une plus grande stabilité chimique que le DNA et le RNA ; il forme des hétéroduplex HNA:RNA et HNA:DNA ainsi que des homoduplex HNA:HNA ; les triphosphates qui lui correspondent (hNTP) sont substrats des DNA polymérases et des rétro-transcriptases.
Nous avons pu établir que de brefs messages génétiques portés par des oligonucléotides HNA pouvaient être transcrits en DNA in vivo. Une souche défective d'Escherichia coli, dont le génome comporte une délétion du gène thyA spécifiant la thymidylate synthase, prolifère seulement si de la thymidine lui est apportée dans son milieu de culture, et ne donne jamais lieu à la réversion spontanée de cette exigence nutritionnelle. Il fut testé si un segment génétique de 18 bases du gène thyA, codant le site actif de l'enzyme et comportant un nombre variable d'hexitol-nucléotides, une fois ligaturé à une construction plasmidique codant le reste du gène thyA, pouvait restaurer l'intégrité de ce gène dans la souche défective. Des colonies capables de proliférer sans apport de thymidine ont été obtenues en ligaturant des oligonucléotides mosaı̈ques contenant un, trois ou six hexitol-nucléotides, avec des fréquences décroissant respectivement dans le rapport 1, 0,4 et 0,005 comparées à la fréquence obtenue avec l'oligonucléotide servant de contrôle et constitué de 18 désoxyribo-nucléotides.
La polymérisation de DNA en réponse à des matrices de HNA fut confirmée in vitro, en utilisant le fragment de la DNA polymérase I d'Escherichia coli dépourvu des activités 5′- et 3′-exonucléase (PolA∗), d'une part, et, d'autre part, la DNA polymérase de Thermus aquaticus (TaqP). Il fut montré que jusqu'à quatre monomères thymidine-triphosphate (dTTP) peuvent s'incorporer en réponse à une matrice penta-hexitol-nucléotide (hA)5. La discrimination de dTTP par rapport à dCTP, dGTP et dATP en réponse à hA dans le brin matrice fut également établie.
Cette propriété de transcription de HNA en DNA par TaqP a été mise en œuvre dans des expériences de PCR à l'aide d'amorces oligonucléotidiques, l'une homogène, constituée de DNA, l'autre mosaı̈que DNA/HNA, comprenant une succession interne d'un nombre variable d'hexitol-nucléotides. Des produits d'amplification furent détectés avec des amorces contenant un et trois hexitol-nucléotides, mais pas avec six hexitol-nucléotides.
Enfin, la démonstration fut apportée que la biosynthèse d'une séquence de HNA par incorporation d'hexitol-nucléotides pouvait être commandée par une séquence de HNA complémentaire utilisée comme matrice sous l'action des DNA polymérases PolA∗ ou TaqP. Jusqu'à trois incorporations successives de hTTP en réponse à une matrice penta-hexitol-nucléotide (hA)5 ont été observées.
Ces résultats reproductibles, qui furent obtenus à l'aide de versions naturelles des DNA polymérases, révèlent un potentiel biosynthétique dans les activités catalytiques marginales de ces enzymes, fût-il restreint à la transmission de courts messages. Ils augurent favorablement de la mobilisation du HNA ex vivo et in vivo, une fois que des versions spécialisées des polymérases auront été obtenues par évolution dirigée. Dans la perspective ex vivo, ces résultats offrent l'ébauche de l'amplification PCR du HNA et de la sélection d'aptamères HNA. Dans la perspective in vivo, ces résultats apportent une première démonstration de faisabilité pour une enclave génétique propageant des messages composés des quatre bases canoniques A, C, G, T et portés par une catégorie supplémentaire de polynucléotide, en d'autres termes des biopolymères informationnels débordant le « dogme central » de la biologie moléculaire : DNA → RNA → protéine.
Nous comptons poursuivre l'exploration des motifs voisins du 1,5-anhydro-hexitol, dans le but d'identifier lesquels pourraient être dérivés du métabolisme des espèces naturelles, ou, au contraire, lesquels resteraient inaccessibles par évolution du métabolisme, et donc seraient moins susceptibles d'être disséminés chez les espèces naturelles.
1 Introduction
The propagation of genetic messages by nucleic acids has been found to involve the pairing of purine and pyrimidine bases attached to a backbone polymer of phospho-ribose or phospho-deoxyribose in all known species. In addition to the four canonical monomers of DNA (dATP, dCTP, dGTP, dTTP) and the four canonical monomers of RNA (ATP, CTP, GTP, UTP), several different alterations of the pyrimidine and purine bases have been reported to occur in the deoxynucleoside triphosphates used as DNA precursors by certain bacterial viruses [1]. By contrast, no backbone deviating from ribose or deoxyribose has thus far been found to be incorporated during polymerisation of natural nucleic acids.
A vast body of data gathered during the elaboration of antisense oligonucleotides [2,3] and the exploration of putative evolutionary precursors of polynucleotides [4], however, establish that polynucleotide analogues constructed with alternative backbones can form base-paired duplexes as stable as or even more stable than DNA and RNA. Even though such polynucleotides have not yet been shown to replicate accurately, these findings strongly suggest that there are numerous backbone motifs that could fulfil the functions of nucleic acids which were not adopted during the evolution of species, yet might be acquired through appropriate reprogramming of living cells. In this context one notes that mutagenic oligonucleotides whose phosphodiester or deoxyribose motifs were modified with sulphur or methyl groups could transfer genetic information [5].
We set out to propagate certain hereditary messages of a microbial cell in the form of sequences of the four bases A, C, G and T (or their respective pairing equivalents 2,6-diaminopurine, 5-methylcytosine, hypoxanthine and uracil, or derivatives thereof) linked to a motif other than deoxyribose or ribose, following the smallest number of genetic steps. In addition to the basic interest that the proliferation of a third type of nucleic acid in vivo would represent in itself, it could also lead to numerous advances in applied domains. One could thus envision the bulk production of functionalized nucleic acids for diagnostic or therapeutic purposes for instance, as well as the establishment of genetic enclaves, i.e. chemically remodelled polynucleotides (heterodox episomes) whose mutability could be tuned to accelerate evolution of certain messages while preserving the hereditary stability of the host DNA genome.
One surmises that the chemical nature of the backbone for such a genetic enclave should satisfy the following trade-off: if it resembled deoxyribose and ribose too closely, it would likely be difficult to prevent interference of the novel nucleic acid monomers with DNA and RNA biosynthesis; if it differed too much from deoxyribose and ribose, it might prove difficult to recruit polymerases and other replication enzymes which could process such heterodox polynucleotides and their monomers.
Of the various motifs elaborated in the course of antisense research, hexitol nucleic acid (HNA) appears to be a promising candidate for in vivo propagation. HNA is built up from nucleobases and a hexitol motif closely mimicking deoxyribose (Fig. 1) [6,7]. We have demonstrated previously that HNA oligonucleotides form very stable self-complementary duplexes as well as sequence-selective heteroduplexes with DNA and RNA [8,9]. In a parallel study, we found that various DNA polymerases could synthesize short HNA stretches of up to six hexitol nucleotides in response to a DNA template [10]. Furthermore, HNA was shown to act as a better template than DNA or RNA for the non-enzymatic condensation of oligonucleotides [11].
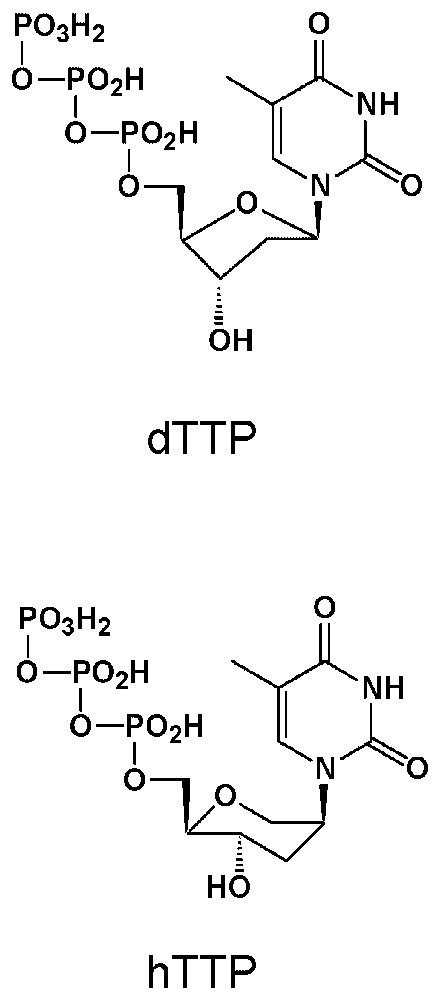
Compared structures of the DNA precursor dTTP and its HNA analogue hTTP.
The results of the investigations that we conducted to assess the feasibility of a HNA-based genetic enclave in E. coli are reported here. It is shown that hexitol oligonucleotides have the capability of instructing the polymerisation of short DNA stretches in vivo, as well as of short HNA stretches in vitro using natural versions of DNA polymerases from E. coli and from Thermus aquaticus.
2 Materials and methods
2.1 General
Ultra-pure dNTPs were purchased from Amersham Pharmacia Biotech. The exonuclease-deficient Klenow fragment of E. coli DNA polymerase I (PolA∗) was purchased from New England Biolabs, Taq DNA polymerase (TaqP) was from Roche Boerhinger.
2.2 Synthesis of hTTP
1,5-Anhydro-2,3-dideoxy-2-(thymin-1-yl)-D-arabino-hexitol (hT) [12] was 5′-phosphorylated according to Tener's method [13]. The monophosphate was then activated as the morpholidate derivative and converted to the corresponding triphosphate by condensation with pyrophosphate according to reported procedures [14]. Crude hTTP was purified by DEAE-cellulose column chromatography (HCO3− form, gradient from 0 to 0.8 M triethylammonium bicarbonate). For DNA polymerase reactions, hTTP was further purified by reverse phase HPLC (C18 column). Triethylammonium salt of hTTP was finally converted to the sodium salt by passing through a Dowex 50W-X8 (Na+ form) column. All intermediates were characterized by NMR spectroscopy. Purity of hTTP was assessed by 31P NMR spectroscopy and mass spectrometry. 31P NMR (D2O) δ: −20.18 (t, Pβ); −9.60 (s, Pα); −4,58 (s, Pγ).
2.3 Oligonucleotides containing hA residues
The phosphoramidite HNA building blocks were synthesized as described before [15]. The oligonucleotides containing several hexitol motifs were synthesized according to previously reported procedures [9] and the correct mass was obtained as determined by negative-ion electrospray mass spectrometry.
2.4 In vitro replication by DNA polymerase
The primer (P7: 5′-CAGGAAACAGCTATGAC-3′) was labelled at its 5′-end with [γ32P]-ATP (4500 Ci mmol−1) and T4 polynucleotide kinase (10 U μl−1). Primer-template duplexes (1:1.5 ratio) were annealed in the kinase buffer by heating at 75 °C for 15 min and cooling slowly to room temperature for 1 h. The elongation reaction mixture contained the corresponding 2× polymerase buffer (10 mM Tris-HCl, 5 mM MgCl2, 7.5 mM DTT (pH 7.5) for PolA∗; 10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl (pH 9), 1% Triton for TaqP), the annealed primer-template duplex (200 nM) and the enzyme at the concentrations indicated in the legends of Fig. 3. The reactions were initiated with the addition of an equal volume of a 2× dNTP solution, incubated for 3 min at 37 °C with PolA∗ or for 15 min at 48 °C with TaqP, and quenched by addition of a gel loading buffer (0.2% bromophenol blue, 0.2% xylene cyanol FF, 80% formamide). The samples were heated at 75 °C for 2 min and loaded onto a denaturing polyacrylamide gel (20%, 7M urea). After electrophoresis, the reaction products were visualized by autoradiography or using a PhosphorImager (Molecular Dynamics).
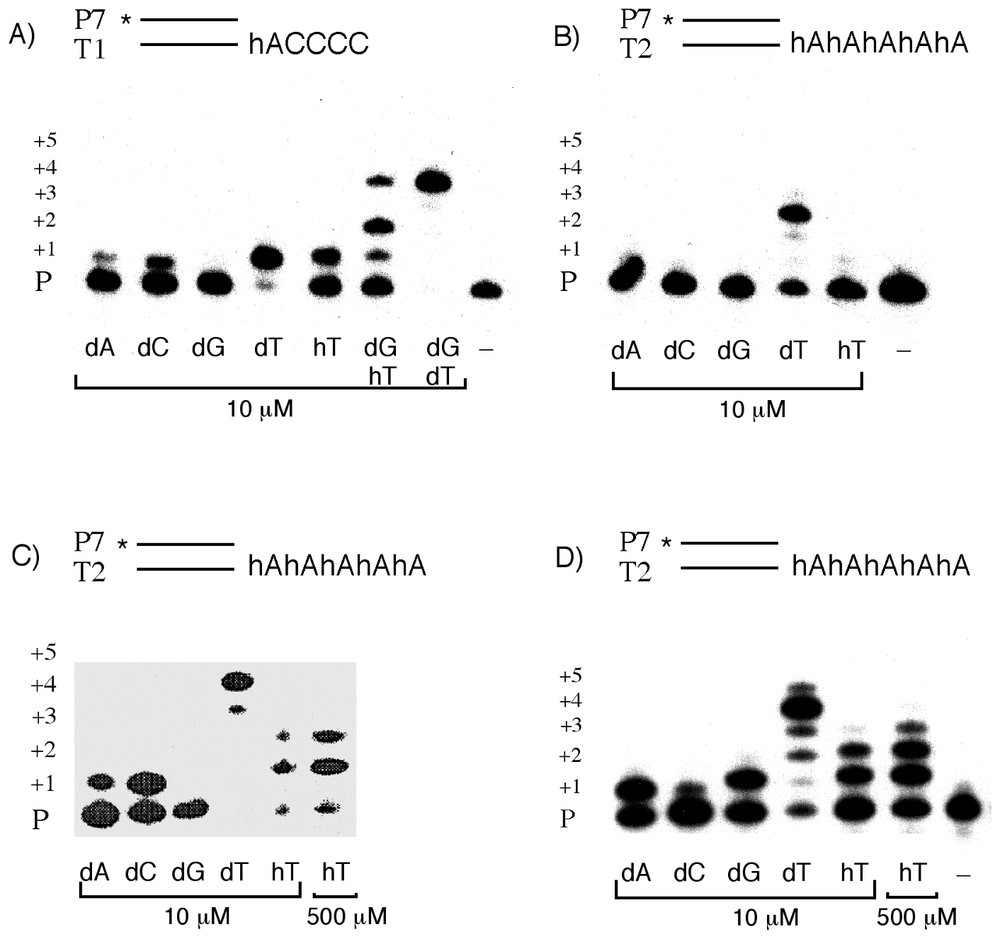
In vitro copying reactions of HNA templates by PolA∗ and TaqP DNA polymerases. Enzymatic condensation of hTTP into hybrids P7:T1 or P7:T2 using templates containing one or five hexitol motifs. PolA∗ was incubated at 0.03 U μl−1 with 100 nM primer:template duplex (A, B) or at 0.05 U μl−1 with 80 nM primer:template duplex (C) in the presence of dNTPs or hTTP as indicated. TaqP was incubated at 0.25 U μl−1 with 100 nM duplex in the presence of dNTPs as indicated (D). Products after incubation for 3 min at 37 °C using PolA∗ and for 15 min at 48 °C using TaqP were analysed by PAGE. Control experiments (incubation without dNTPs) are shown on the right of the panels.
2.5 PCR amplification
For the PCR amplification experiments, 25 pmol of forward primer, 25 pmol of oligonucleotide (P2, P3, P4, P5 or P6 listed in Table 1), 1 ng of pTZ18thyA DNA template (see below) and 200 μM of each dNTP were mixed in reaction buffer (Roche) and Taq DNA polymerase (1 U) was added in a 50 μl final volume. The cycling parameters were: 5 min, 95 °C; 25 × (30 s, 95 °C; 30 s, 35 °C < T<55 °C; 30 s, 72 °C); 10 min, 72 °C. The samples (5 μl) were analysed by electrophoresis on a 1% agarose gel.
Transmission of hexitol-supported messages to restore thymidylate synthase activity in E. coli in a gapped heteroduplex ligation assay
| Sequence of 5′-phosphorylated oligonucleotide | Number of hexitol motifs | Thy+(a) | Bla+(b) | Thy+/Bla+(c) | |
| none | 0 | 0 | 19 592 | 0 | |
| P1 | CTAGCGCCG---CATGCA | 0 | 0 | 18 936 | 0 |
| P2 | CTAGCGCCGTGCCATGCA | 0 | 2967 | 39 095 | 0.0760 |
| P3 | CTAGCGCCG GCCATGCA | 1 | 1526 | 21 267 | 0.0717 |
| P4 | CTAGCGCCG CATGCA | 3 | 637 | 27 002 | 0.0236 |
| P5 | CTAGCG CATGCA | 6 | 83 | 49 651 | 0.0017 |
| P6 | CTAGCG CATGCA | 6 | 9 | 33 124 | 0.0003 |
(a) Bases of hexitol nucleotides are underlined in the sequence.
(b) Thy+ refers to the number of thymidine-prototrophic colonies.
(c) Bla+ refers to the total number of transformant colonies (Thy+ and Thy−).
2.6 Plasmid construction
The pTZ18thyA plasmid was obtained by site directed mutagenesis of pTS0 (pTZ 18R containing the E. coli thyA gene at the HindIII site [16] to introduce a NheI restriction site at positions 429 of the coding sequence (unpublished data). The pTZ18thyA plasmid was digested with the NheI and NsiI restriction enzymes, then ligated to a NheI-NsiI 1.1 kb DNA fragment (spc+) conferring resistance to spectinomycin and streptomycin from pHP45 [17] to produce the plasmid pAK1. The pAK1 plasmid was digested with NheI and NsiI, purified by electrophoresis on a 1% agarose gel and ligated to the oligonucleotide 5′-pCTAGTGCA-3′ to produce pAK2. The pAK2 plasmid bears a deletion of 10 nucleotides and cannot code for an active thymidylate synthase.
2.7 Heteroduplex formation
Equimolar amount (50 ng) of pAK1, cleaved with NheI and NsiI and purified on a 0.7% agarose gel, and pAK2, linearized with EcoRI, were mixed in a 10 mM Tris-HCl (pH 7.5), 100 mM NaCl buffer, then denatured for 5 min at 85 °C, and slowly cooled to room temperature over 1 hour. The heteroduplexes formed were desalted through a Microcon column (Millipore).
2.8 Ligation and transformation
The 18-mer oligonucleotides used were listed in Table 1. Each oligonucleotide (2 pmol) was hybridised with the heteroduplexes described above (0.02 pmol) in annealing buffer (50 mM Tris-HCl, 10 mM MgCl2, 10 mM DTT, 25 μg ml−1 BSA pH 8.0) by heat denaturation for 5 min at 85 °C, followed by slow cooling to room temperature over 1 h. Ligation was performed in the annealing buffer supplemented with 1 mM ATP and 5U of T4 DNA ligase by incubating the mixtures at 16 °C overnight. An aliquot of ligation mixtures was used to transform CaCl2 competent cells of the strain β1308 (ΔthyA::erm) [18]. After 30 min of incubation at 4 °C, the cells were shocked for 5 min at 37 °C and 1 ml of Muller–Hinton (MH) medium, containing 0.3 mM thymidine was added. After a 1-h incubation at 37 °C with agitation, the cells were centrifuged and washed twice in the same medium without thymidine. Part (1/10) of the transformation mixture was plated on MH agar plates containing thymidine (0.3 mM) and carbenicillin (100 μg ml−1) and the remaining 9/10 on MH agar plates.
3 Results and discussion
3.1 HNA-dependent DNA polymerisation in vivo
A selection scheme was designed to assay transmission of genetic messages from HNA oligonucleotides to a DNA replicon by the replication enzymes in a living bacterium. It consisted of transforming E. coli cells with plasmids bearing a HNA:DNA heteroduplex encoding a functional and selectable gene, such that propagation of that gene would require utilization of the HNA stretch as a template (Fig. 2A). Synthetic 5′-phosphorylated 18-mer mosaic HNA/DNA oligonucleotides encoding the active site region surrounding the catalytic Cys146 of E. coli thymidylate synthase were ligated to a gapped vector carrying an inactive allele of the E. coli thyA gene that precisely lacked the codons for that catalytic region (Fig. 2B). Since active thymidylate synthase is absolutely required for growth of E. coli in nutrient media devoid of thymine or thymidine, this system was expected to provide a tight selection for HNA copying into DNA in vivo.
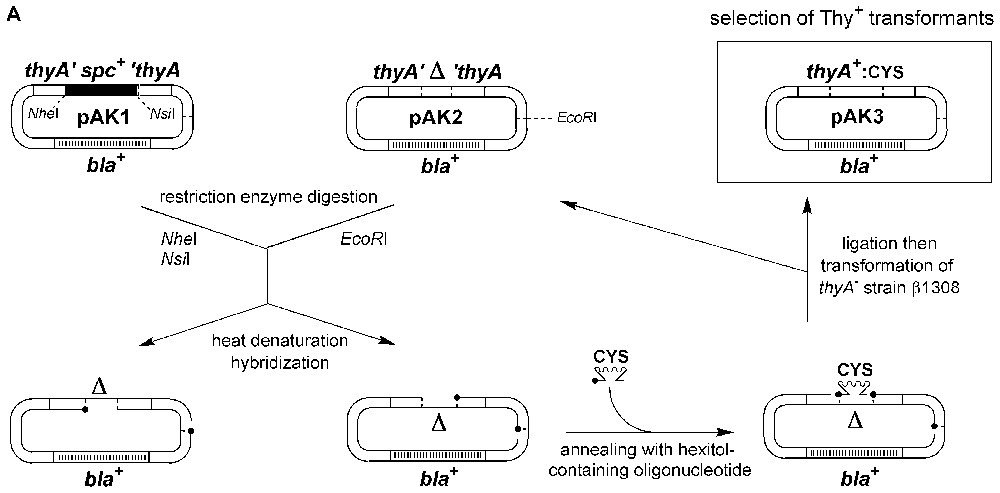
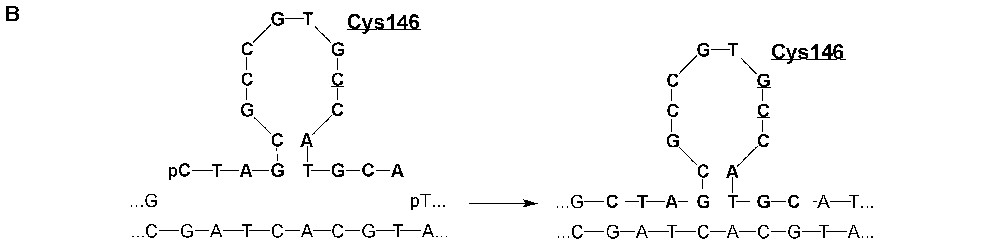
Transmission of hexitol-supported messages to restore thymidylate synthase activity in E. coli using a gapped heteroduplex ligation assay. A. Both pAK1 and pAK2 plasmids are ColE1 derivatives containing the bla gene (beta-lactamase) conferring ampicillin resistance. pAK1 carries the E. coli gene for thymidylate synthase (thyA) inactivated by replacing the codons 143–148 with a spectinomycin resistance gene (spc) inserted between the NheI and NsiI restriction sites. pAK2 carries the thyA gene inactivated by deleting the codons 143–148. Following restriction enzyme digestion, heat denaturation and hybridisation, the resulting heteroduplexes were ligated with synthetic 5′-phosphorylated mosaic HNA/DNA 18-mers encoding the active site region surrounding the catalytic residue Cys146 (CYS) of ThyA. After transformation of the Thy− auxotrophic strain β1308 (ΔthyA::erm) with the ligation products, Thy+ prototrophs were selected for growth on medium devoid of thymidine. B. Close-up of the ligation reaction restoring the integrity of the thyA gene sequence. Notice that neither the sequence encoding the catalytic site of thymidylate synthase around Cys146 nor its complement occurs in a continuous DNA stretch in any component of the ligation/transformation assay and that such a continuous DNA stretch can only be generated by copying the HNA stretch.
The length of contiguous hexitol nucleotides in the mosaic oligonucleotides was set as the experimental variable. Prototrophic transformants (i.e. capable of growth without added thymidine) of the E. coli strain β1308 deleted for the thyA gene could be reproducibly obtained by ligating the carrier vector to mosaic oligonucleotides with 1-, 3- and 6-unit long hexitol nucleotide stretches (Table 1). As expected, ligation of the 15-mer DNA oligonucleotide lacking the Cys146 codon (P1) did not yield any prototrophic transformants. Omitting oligonucleotides, carrier vector, or ligase in heteroduplex assembly also yielded no prototrophic transformants. Similar amounts of thymidine prototrophic transformants were obtained for a mosaic oligonucleotide bearing one hexitol (P3) and the control 18-mer DNA oligonucleotide (P2).
A 2.5-fold drop in the yield of prototrophic transformants resulted from extending the hexitol stretch from one to three nucleotides, and a close to 200-fold drop from extending the hexitol stretch from one to six nucleotides. Sequencing of the plasmids conferring thymidine prototrophy demonstrated that the message conveyed in hexitol stretches was correctly copied into DNA in all cases. In particular, the thyA sequences resulting from ligation of the mosaic oligonucleotides P5 and P6, both of which encoding the wild-type amino acids of the ThyA catalytic region but containing two silent mutations, were found to match exactly the 6-long hexitol stretches from which they respectively originated. This result indicated accurate copying of hexitol templates. Thus, the decrease in the yield of active thyA genes among clones transformed using hexitol templates was probably caused by inefficient rather than erroneous copying by E. coli DNA polymerases. The inefficiency of ligating mosaic HNA/DNA oligonucleotides to carrier vectors cannot be excluded, however. The identity of the DNA polymerases transcribing HNA messages into DNA in E. coli is currently being investigated.
3.2 HNA-dependent DNA polymerisation in vitro
DNA synthesis in response to hexitol-containing templates was examined in vitro by primer elongation reactions catalyzed by the exonuclease-deficient Klenow fragment of DNA polymerase I (PolA∗). Two duplexes were designed in which the templates contain either one hA nucleotide followed by four dC nucleotides (T1) or a stretch of five hA nucleotides (T2). The integrity of the synthesized templates was verified by mass spectrometry. Elongation products obtained with T1 in the presence of each of the four canonical nucleoside triphosphates as well as in the presence of dTTP and dGTP are shown in Fig. 3A. Under experimental conditions which minimize the frequency of base misincorporations (see Section 2 Materials and methods for details), dTMP was best incorporated opposite the hA nucleotide leading to nearly complete elongation of the primer. There was only minor incorporation of dCMP and dAMP and no detectable incorporation of dGMP. With a mixture of dTTP and dGTP, all the primer was elongated and a single product was formed which corresponded to the primer elongated by four nucleotides. These results indicated that PolA∗ is able to copy a base linked to the hexitol motif with a strong preference for the complementary base and that once the hA:dT nucleotide pair is formed, the enzyme can readily copy several dCMP residues of the template. Applying the same experimental conditions to the template T2, there was no detectable incorporation of dAMP, dCMP or dGMP whereas polymerization of dTMP was quite efficient with three residues incorporated opposite the hexitol motifs. The base specificity was thus again preserved (Fig. 3B). At higher enzyme/template-primer ratio (2-fold) or higher dTTP concentration (500 μM), all the primer was utilized and a product was formed by condensing four dTMP residues (Fig. 3C).
3.3 HNA-dependent HNA polymerisation in vitro
Our observation that PolA∗ could copy hexitol containing templates into DNA led us to test whether the polymerase was also endowed with HNA-dependent HNA polymerase activity. The hexitol thymidine triphosphate (hTTP) was obtained following a synthetic route that excluded the concomitant formation of dTTP, as confirmed by spectroscopic characteristics (see Section 2 Materials and methods). We examined the condensation of hTTP with templates containing one or several adenine hexitol motifs. With the template containing one hA nucleotide (T1), the incorporation of hTMP was not as efficient as the incorporation of dTMP but much better than the incorporation of dCMP or dAMP, with about half of the primer elongated (Fig. 3A). In the presence of both hTTP and dGTP, again about half of the primer was elongated. The major product corresponded to a primer elongated by two nucleotides. There were two other products with one and four nucleotides incorporated (Fig. 3A). With the five-long hexitol stretch (T2), incorporation of hTMP was not detectable (Fig. 3B). However, when the enzyme/template-primer ratio was doubled or the concentration of hTTP raised to 500 μM, elongation products were obtained with one and two hTMP incorporated (Fig. 3C). Polymerization products obtained with T2 by the action of TaqP in place of PolA∗ in the presence of each of the four canonical nucleotides and hTTP are shown in Fig. 3D. As with PolA∗, hTMP was incorporated opposite the hA nucleotide, albeit less efficiently than dTMP. At higher hTTP concentration (500 μM), the formation of the product with three incorporated hTMPs was increased.
3.4 PCR amplification with a mosaic DNA/HNA primer
We also tested the introduction of hexitol motifs in polymerase chain reactions, elaborating on the fact that primer elongation with deoxynucleotides was catalyzed by TaqP in response to hexitol motifs located in a template (see Fig. 4). We thus attempted to measure the templating efficiency of hexitol-containing primers by the production of nucleic acid through PCR amplification. The oligonucleotides designed for in vivo transmission of HNA-carried messages in E. coli (Table 1) were used for this purpose, with the E. coli thyA gene as the DNA template (590 bp amplification product). Mosaic oligonucleotides (18-mers) differing by the length of the hexitol stretch were used as upstream primers, and a standard DNA oligonucleotide (M13 forward primer) as the downstream primer. The annealing temperature for amplifications was systematically tested for each couple of primers (see Section 2 Materials and methods). When the upstream primer contained one hexitol (P3), the amplification was similar to that obtained with the control DNA upstream primer (P2) (Fig. 4A and B). With a stretch of three hexitol motifs in the upstream primer (P4), the quantity of nucleic acid product was slightly reduced (Fig. 4C). No product could be detected with upstream primers containing six hexitol motifs (Fig. 4D). This could be explained either by the instability of the duplex formed with DNA or by the inability of the TaqP to copy long HNA stretches. Overall, these results were in agreement with those obtained both in vivo and in the primer extension experiments.
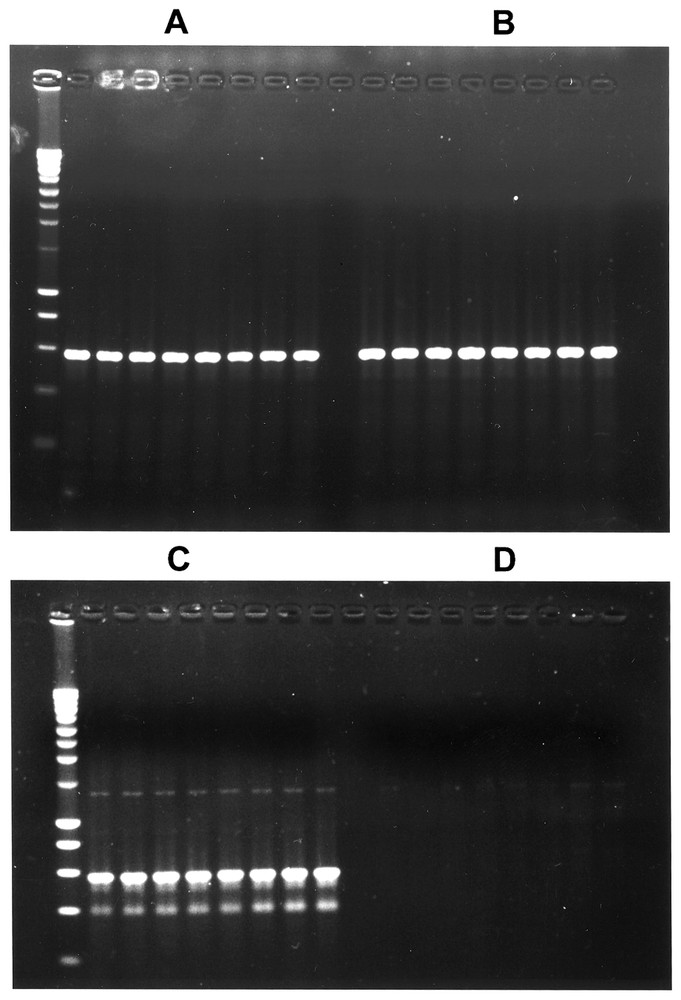
PCR amplification using mosaic DNA/HNA oligonucleotides as primers. The M13 forward primer was used in all amplification reactions in combination with a variable mosaic oligonucleotide differing by the number of hexitol residues, and consisting of oligonucleotide P2 (no hexitol, A), P3 (one hexitol, B), P4 (three hexitols, C), and P5 (six hexitols, D). The temperature of annealing increased from 35 °C (left lane) to 55 °C (right lane) by increments of 2.8 °C in each panel. Products were separated by 1% agarose gel electrophoresis. The standard ladder (Eurogentec) stands on the left side of panels A and C.
4 Conclusion
The significance of the results reported here should be considered within the long-term perspective of propagating a novel type of genetic material in a microbial cell. The transmission of a genetic message encoding two codons carried on a hexitol hexamer could be reproducibly demonstrated in E. coli cells. Although the quantity of information thus conveyed may appear modest, it was obtained with natural polymerase activities. Moreover, it was unexpected that the common enzymes PolA∗ and TaqP would be able to accommodate substitution of deoxyribose with hexitol in the nucleoside triphosphate and polynucleotide alcohol substrates, as well as in the template co-catalyst. Mutation studies of polymerase active sites have shown that amino acid substitutions can lead to altered nucleoside triphosphate recognition [19]. It can thus be reasonably expected that a limited number of mutations in DNA polymerases will make it possible to replicate longer HNA duplexes and that the fixation of appropriate mutations in TaqP will allow PCR amplification of HNA duplexes, and possibly the evolution in vitro of HNA aptamers. The genetic screen elaborated for this study could be used in the future with longer HNA messages in order to select for polymerase mutants with higher efficiency toward HNA templates and hexitol monomers.
Evolving a bona fide HNA-dependent HNA polymerase will presumably not be sufficient to propagate a HNA-based genetic enclave, since other enzymes such as HNA-binding proteins or HNA ligases will also probably have to be adapted to HNA substrates. In addition, a functional requirement will be that the HNA encoded message, however short, should be mandatory for the survival of the host cell. In this regard, the selection of an HNA-dependent RNA polymerase might also have to be considered. Alternatively, translation of HNA by ribosomes [20] to produce a short vital peptide, or catalysis of an essential metabolic step by a HNA aptamer might have to be implemented in vivo.
The fact that HNA oligonucleotides can be copied by natural polymerase enzymes incites us to explore heterodox polynucleotides further deviating from DNA and RNA. A future goal could be to elaborate a backbone motif that is rejected under the form of nucleoside triphosphate precursors by the polymerases of the host cell, and then to evolve specialized versions of polymerases for establishing a genetic enclave whose biosynthesis would not interfere with DNA and RNA metabolism. A phosphosugar motif accessible from metabolic precursors present in the host through a limited number of steps might be defined as an additional criterion for that search, in order to achieve biosynthetic autonomy of the genetic enclave.
Alternatively, one might seek to enforce the nutritional containment of organisms harbouring a genetic enclave and to disable the dissemination thereof, by elaborating backbones ever further deviating from phosphopentoses, hence ever less accessible from extant metabolic routes. Such genetic enclaves could be propagated only if chemical precursors of heterodox polynucleotides were supplied to their hosts as synthetic nutrients unavailable from natural food chains.
Acknowledgements
We are grateful to Susan Cure and Catherine Papanicolaou for improving the manuscript and to Madeleine Bouzon for discussion.


