1 Introduction
Good systematics and taxonomy are desirable for understanding the biogeographical patterns of species diversity. Of all the marine living organisms of the Indo-West Pacific, Teleostean fish probably are among the best-described, even though their systematics and taxonomy still need considerable research effort [1,2].
The fish family Siganidae (‘rabbitfishes’) comprises a single genus, Siganus, with 28 nominal species validated so far [3,4]. Species richness in Siganidae decreases from the central Indo-West Pacific region, where a majority of the species are thought to originate from, to the periphery [3,5]. This pattern has been observed for other groups of marine organisms [6]. Therefore, Siganidae constitute an interesting model-group to study speciation processes in the Indo-West Pacific, but their taxonomy first requires a precise re-evaluation. Remarkable homogeneity in those characters that usually allow one to diagnose species, such as numbers of fin spines and soft rays, tooth count and tooth shape and size, and broad across-species overlap in other characters, such as gill-raker and lateral-line scale counts [3,4] have earlier led to taxonomic confusion in Siganidae. The present taxonomy of Siganidae, based on Woodland's (1990) extensive revision [3] relies on colour patterns and gross morphological features, such as body proportions and relative length of snout [3,4]. Apart from an allozyme survey describing the genetic relationships among 10 species of the family, 9 of which were sampled from a localised region of the tropical West-Pacific [7], little emphasis has been placed so far on validating the current classification using genetic markers.
Fish of the family Siganidae occur in the coastal waters of the tropical Indo-Pacific. Two species occurring in the Red Sea, S. luridus and S. rivulatus, have recently expanded their area of distribution into the Mediterranean across the Suez Canal [8]. The geographic distributions of rabbitfish range from the entire tropical Indo-West Pacific except the easternmost Pacific islands to regional, presumably, in part, as a consequence of differences in the dispersal potential of juveniles [3]. A proportion of species are originally endemic to restricted geographic areas such as the enclosed Red Sea (S. rivulatus) and Andaman Sea (S. magnificus), and isolated groups of islands or reefs (Maldives: S. puelloides; scattered reefs off Western Australia: S. trispilos; Fiji: S. uspi; Tonga: S. niger) [3]. One species (S. unimaculatus) has an antitropical distribution [3]. There might be a second example of antitropical distribution in Siganus: S. woodlandi [4] has so far been reported from New Caledonia only, but the sedaka-hana-aigo, an undescribed rabbitfish from Japan “that appears to be either S. woodlandi or a very close relative” ([4], and references therein) might be its antitropical counterpart. Pairs of presumed sibling species have been reported [3]. The latter have an essentially Indian/West Pacific distribution with some overlap in the central Indo-Malay region [3]. Among these, morphologically intermediate individuals thought to be hybrids between S. doliatus and S. virgatus have been reported [3,9], and hybrids between S. guttatus and S. lineatus are similarly expected [9].
Here, we analysed the nucleotide sequences of a fragment of the cytochrome b gene in 16 Siganidae species sampled across the Indo-West Pacific. Our objectives were: (1) to evaluate the potential of the cytochrome b gene as a species-specific marker (a DNA-barcode) in Siganidae; (2) to quantify genetic differences between sibling species with parapatric distribution; (3) to estimate the degree of genetic differentiation among populations within a few species differing by the width of their geographic distribution.
2 Materials and methods
Two hundred and sixty-two rabbitfish belonging to 15 nominal species, and 22 unidentified rabbitfish larvae were collected from seven locations in the Indo-West Pacific and the Mediterranean (Table 1). The fish were obtained from fishermen (all samples from Ouvéa, Makassar and Truk, and most samples from the southern lagoon of New Caledonia), or as side-samples of ecological surveys (the S. spinus sample of Moorea, and some samples from the southern lagoon of New Caledonia including all larvae, which were collected using light traps [11]). Voucher-specimens for all Siganidae species collected in the southern lagoon of New Caledonia and in Moorea were deposited at the Museum national d'histoire naturelle, Paris (MNHN 2006-1525 to 1537). Muscle tissue samples of S. luridus and S. rivulatus, preserved frozen at Station méditerranéenne de l'environnement littoral, Sète, were sent to us by F. Bonhomme. A piece of muscle tissue of a S. lineatus specimen from the Maldives, deposited at the Bishop Museum, Honolulu (BPBM 40390), was donated to us by J.E. Randall. For one more species (S. virgatus) we only had at our disposal cytochrome b gene sequences available from the GenBank sequence database (http://www.ncbi.nih.gov/Genbank: accession nos. AY190546, AY190549, and AY190550). The samples from the Makassar Strait region (South Sulawesi, Indonesia) included a group of nine individuals whose coloration patterns ranged from typical S. guttatus to typical S. lineatus [3], through a range of intermediate patterns (for seven individuals), indicating possible introgression between the two species.
Samples of Siganidae and Acanthuridae analysed for the present survey
| Species, | Sampling date | N | Collector |
| Sampling location | |||
| Acanthurus blochii | |||
| Southern lagoon, New Caledonia | Apr. 2006 | 2 | P. Borsa |
| Siganus argenteus | |||
| Southern lagoon, New Caledonia | June–July 2005; Mar. 2006 | 24 | P. Borsa, J.-L. Justine, P. Villard |
| Ouvea lagoon, Loyalty Islands | Feb. 2006 | 24 | M. Leopold |
| Truk, Caroline Islands, FSM | Apr. 2006 | 23 | L. Vigliola |
| S. corallinus | |||
| Southern lagoon, New Caledonia | June–July 2005 | 2 | P. Borsa |
| S. doliatus | |||
| Southern lagoon, New Caledonia | June 2005–Apr. 2006 | 9 | P. Borsa |
| Truk, Caroline Islands, FSM | Apr. 2006 | 4 | L. Vigliola |
| S. fuscescens | |||
| Southern lagoon, New Caledonia | June 2005–Feb. 2006 | 10 | P. Borsa, J.-L. Justine |
| Southern lagoon, New Caledonia | 2005 | 3 | C. Mellin |
| Philippines | < May 2002 | 4 | [10] |
| S. guttatus | |||
| Makassar, Sulawesi, Indonesia | Sep. 2005 | 11 | P. Borsa |
| S. javus | |||
| Makassar, Sulawesi, Indonesia | Sep. 2005 | 4 | P. Borsa |
| S. lineatus | |||
| New Caledonia | Apr. 2002–Mar. 2006 | 13 | P. Borsa |
| Makassar, Sulawesi, Indonesia | Sep. 2005 | 5 | P. Borsa |
| Addu Atoll, Maldives | Aug. 2005 | 1 | R.C. Anderson |
| Philippines | < May 2002 | 1 | [10] 1 |
| S. luridus | |||
| Lattaqiah, Syria | 2002 | 7 | M. Hassan |
| S. puellus | |||
| Southern lagoon, New Caledonia | June–July 2005 | 12 | P. Borsa |
| Truk, Caroline Islands, FSM | Apr. 2006 | 9 | L. Vigliola |
| S. punctatus | |||
| Southern lagoon, New Caledonia | June 2005; Mar. 2006 | 21 | P. Borsa, J.-L. Justine |
| Makassar, Sulawesi, Indonesia | Sep. 2005 | 10 | P. Borsa |
| Ouvea lagoon, Loyalty Islands | Feb. 2006 | 3 | M. Leopold |
| Truk, Caroline Islands, FSM | Apr. 2006 | 12 | L. Vigliola |
| Philippines | < May 2002 | 1 | [10] 2 |
| S. rivulatus | |||
| Lattaqiah, Syria | 2002 | 7 | M. Hassan |
| S. spinus | |||
| Southern lagoon, New Caledonia | Jan. 2003; June 2005 | 1 | P. Borsa |
| Moorea, French Polynesia | Apr. 2006 | 5 | D. Lecchini |
| S. vermiculatus | |||
| Makassar, Sulawesi, Indonesia | Sep. 2005 | 2 | P. Borsa |
| S. virgatus | |||
| Philippines | < May 2002 | 3 | [10] |
| S. vulpinus | |||
| Southern lagoon, New Caledonia | July 2005 | 9 | P. Borsa |
| Truk, Caroline Islands, FSM | Apr. 2006 | 22 | L. Vigliola |
| Philippines | < May 2002 | 1 | [10] |
| S. woodlandi | |||
| Southern lagoon, New Caledonia | June–July 2005 | 9 | P. Borsa |
| Unidentified larvae | |||
| Southern lagoon, New Caledonia | 2004–2005 | 22 | L. Carassou |
DNA was extracted from ethanol-preserved fin clips or muscle fragments using either the standard phenol–chloroform extractions with Phase-Lock GelTM tubes (Eppendorf, Le Pecq, France), or the DNeasy® kit (Qiagen GmbH, Hilden, Germany). Individual DNA extracts were subjected to PCR amplification of a 326-bp fragment of the cytochrome b gene, using a pair of degenerate primers (forward: 5′-TCMGAYATCGCCACAGCCTTCTC-3′; reverse: 5′-AGGAAGTGGAAKGCGAAGAA-3′) designed from the alignment of the cytochrome b gene sequences of Siganidae available in GenBank (accession nos. AY190542 to AY190555). The amplification reactions were done in 96-well plates with each well containing 20 μL reaction mixture (2 μL template DNA, 2.5 mM MgCl2, 0.48 mM dNTP mix, 0.5 μM of each primer and 0.5 polymerase (Promega, Madison WI, USA)) in a RobocyclerTM thermocycler (Stratagene, La Jolla CA, USA). The PCR program consisted of 3 min DNA-denaturation (at 94 °C) followed by 35 cycles of 1 min denaturation (94 °C)/1 min annealing (60 °C)/1 min elongation (72 °C). The PCR products were shipped to GATC Biotech (Konstanz, Germany), or Macrogen (Seoul, South Korea) for nucleotide sequencing and the sequences were subsequently retrieved from the company's Internet website.
All nucleotide sequences, which were deposited in GenBank (accession numbers DQ898027 to DQ898094), were aligned, together with their homologues from the Philippines (GenBank AY190543 to AY190556), using BioEdit [12]. The phylogenetic relationships among species in the genus Siganus were inferred from the matrix of haplotype sequences using three different algorithms: neighbour-joining (NJ [13]), maximum parsimony (MP [14]), and maximum likelihood (ML [15]). NJ and MP analyses were done using Mega 3.1 [16]. ML analysis was done using Phylip, version 3.65 [17]. Nucleotide distances between haplotypes () were estimated according to Kimura's 2-parameter model of nucleotide substitution ([18], implemented in Mega 3.1). Gaps were treated as deletions in pairwise comparisons of sequences (option ‘Pairwise-Deletion’ in Mega 3.1). Haplotype frequency differences between populations were estimated using , Weir and Cockerham's fixation-index estimator, which apportions the genetic diversity among samples relative to the total diversity [19]. Tests of significance were done by comparing the observed -value with 1000 pseudo- produced by random permutations of haplotypes among samples using Genetix 4.05 [20]. Taking into account the information harboured in the nucleotide sequences of each allele along with allele frequencies, we used analysis of molecular variance (AMOVA [21]) implemented in Arlequin 3.01 [22] to estimate , Excoffier et al.'s (1992) fixation index for nucleotide sequence data [21]. The -estimate produced by AMOVA is equivalent to a multiple-locus where each variable nucleotide site is considered as a different locus [23]. The significance of -estimates was tested using the non-parametric permutation algorithm implemented in Arlequin 3.01.
3 Results
The mitochondrial phylogeny of Siganidae, based on a 300-bp fragment of the cytochrome b gene, displayed clusters of haplotypes or haplogroups atop long branches, generally supported by high bootstrap scores (Fig. 1). In all cases, a haplogroup corresponded to a nominal species, except for the pair S. doliatus/S. virgatus, whose haplotypes formed a single, tight cluster. The homogeneity of S. fuscescens was questionable, as the haplotypes sampled for that species appeared to form two separate haplogroups, distant by . The haplogroup corresponding to S. guttatus was weakly supported and appeared to be nested within a S. guttatus/S. lineatus cluster, whose most external haplotype was that found in S. lineatus from the Maldives. The S. guttatus haplogroup was distant from the Indo-West Pacific (i.e., Makassar Strait, Philippines, New Caledonia) S. lineatus by while the latter were separated from the Maldives S. lineatus haplotype by . S. luridus and S. spinus were separated by a high nucleotide distance ( and the topology of the tree provided little support to the hypothesis that they form sister-clades. S. vulpinus haplotypes formed a sub-clade of a strongly supported clade that also included S. puellus and S. punctatus. S. woodlandi haplotypes formed a monophyletic clade with S. argenteus, distant from it by .
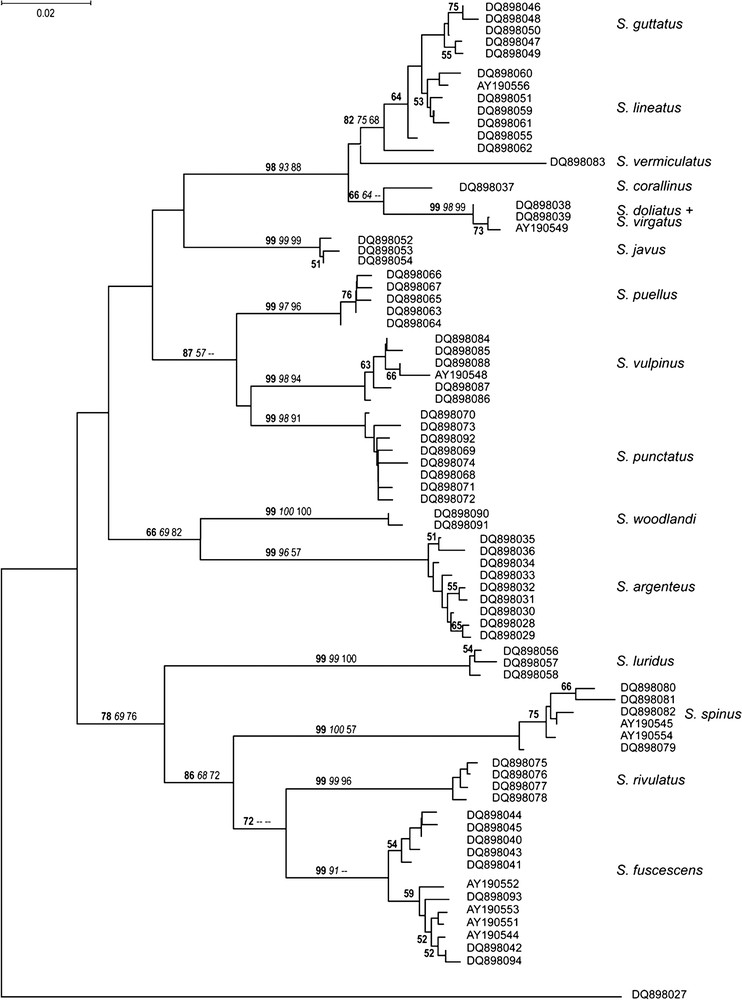
Mitochondrial phylogeny of Siganidae (Siganus spp.). Neighbour-joining tree (Kimura's 2-parameter nucleotide distances [18]; Mega 3.1 [16]) of nucleotide sequences of a 300-bp fragment of the cytochrome b gene (GenBank DQ898028 to DQ898094 and AY190543 to AY190556). Homologous sequence in Acanthurus blochii (Acanthuridae) (GenBank DQ898027) used as outgroup [24]. Numbers at a node are percentage scores (>50%) after bootstrap resampling [25] of nucleotide sites in the matrix of sequences (bold: neighbour-joining, 1000 bootstraps; italics: maximum parsimony (Mega 3.1), 1000 bootstraps; regular: maximum likelihood (Phylip 3.65 [17]), 100 bootstraps; for the sake of clarity, the two latter percentages were figured on the main nodes only). Scale bar: 2% nucleotide divergence estimate. Synonymous sequences: AY190549 = AY190550; DQ898039 = AY190546; DQ898047 = AY190547; DQ898054 = AY190555; DQ898068 = AY190543.
The haplotypes of seven individuals morphologically intermediate between S. guttatus and S. lineatus were equally distributed among the two respective haplogroups characteristic of each species. The haplotypes of the 22 unidentified larvae sampled in the southern lagoon of New Caledonia clustered with either S. lineatus or S. fuscescens.
The degree of population differentiation, as estimated by fixation indices at the scale of the Indo-West Pacific, was significant in seven of the eight species tested (Table 2). The high degree of genetic differentiation found among S. fuscescens populations, however, could be misleading in the case where two reproductively isolated populations had been sampled in New Caledonia (see above). Accordingly, we re-estimated the fixation-index values after considering solely the S. fuscescens clade formed by haplotypes DQ898042, DQ898093, AY190545, and AY190551 to AY190553, represented in both New Caledonia and the Philippines, and found () and (). The fixation indices could not be estimated in the other S. fuscescens clade (formed by haplotypes DQ898040, DQ898041, DQ898043 to DQ898045, and DQ898094) since the latter was represented in the New Caledonian sample only.
Siganus spp. mitochondrial haplotype (partial sequence of cytochrome b gene) frequencies by sample. All sequence data were from the present survey, except for 10 Philippines specimens, whose sequences were retrieved from GenBank (details in footnote). Degree of genetic differentiation estimated using Weir and Cockerham's [19,20] and Excoffier et al.'s [21,22]. Nucleotide sites numbered from 1 to 300, with the first site homologous to site no. 14611 in Atlantic salmon, Salmo salar (GenBank AF133701)
| Species, | Polymorphic site | Sample | ||||||
| Haplotype | NC | Ouvéa | Truk | MS | Moorea | |||
| 1222 | ||||||||
| 35792479 | 0.020⁎ | 0.031 | ||||||
| S. argenteus | 61831931 | |||||||
| DQ898028 | CACTGCCT | 12 | 14 | 6 | – | – | ||
| DQ898029 | .......C | 0 | 0 | 1 | – | – | ||
| DQ898030 | .......C | 4 | 6 | 13 | – | – | ||
| DQ898031 | ......TC | 0 | 1 | 0 | – | – | ||
| DQ898032 | ....A.TC | 2 | 0 | 0 | – | – | ||
| DQ898033 | .G.....C | 1 | 0 | 0 | – | – | ||
| DQ898034 | .....A.C | 0 | 1 | 0 | – | – | ||
| DQ898035 | ...C...C | 4 | 1 | 3 | – | – | ||
| DQ898036 | T.TC...C | 1 | 1 | 0 | – | – | ||
| 12 | ||||||||
| 55 | 0.352⁎ | 0.291⁎ | ||||||
| S. doliatus | 32 | |||||||
| DQ898038 | TC | 7 | – | 4 | 0 | – | ||
| DQ898039 | .T | 2 | – | 0 | 11 | – | ||
| AY190549 | C. | 0 | – | 0 | 21 | – | ||
| 11111222 | ||||||||
| 2577925668277 | 0.152⁎⁎ | 0.279⁎ | ||||||
| S. fuscescens | 37423339683636 | |||||||
| DQ898040 | AACGTATTCAGGCG | 3 | – | – | 0 | – | ||
| DQ898041 | ......C....... | 11 | – | – | 0 | – | ||
| DQ898042 | .G.A.G.CT..... | 11 | – | – | 0 | – | ||
| DQ898043 | ....C...T..... | 1 | – | – | 0 | – | ||
| DQ898044 | ........T..... | 3 | – | – | 0 | – | ||
| DQ898045 | ...........A.. | 2 | – | – | 0 | – | ||
| DQ898093 | GG...G.CT....A | 1 | – | – | 0 | – | ||
| DQ898094 | ..A.....T..... | 1 | – | – | 0 | – | ||
| AY190544 | .G.A.G.CT.A... | 0 | – | – | 12 | – | ||
| AY190551 | .G...G.CT..... | 0 | – | – | 12 | – | ||
| AY190552 | .G...C.CT...T. | 0 | – | – | 12 | – | ||
| AY190553 | .G...G.CTG.... | 0 | – | – | 12 | – | ||
| 11222 | ||||||||
| 816379 | 0.131⁎ | 0.394⁎⁎⁎ | ||||||
| S. lineatus | 143634 | |||||||
| DQ898051 | TCGTCG | 0 | – | – | 1 | – | ||
| DQ898055 | CT...A | 10 | – | – | 0 | – | ||
| DQ898059 | CT.... | 3 | – | – | 4 | – | ||
| DQ898060 | CT.GTA | 1 | – | – | 0 | – | ||
| DQ898061 | CTT... | 1 | – | – | 0 | – | ||
| AY190556 | CT.G.. | 0 | – | – | 13 | – | ||
| 1112 | ||||||||
| 1882 | 0.101⁎ | 0.161⁎ | ||||||
| S. puellus | 7696 | |||||||
| DQ898063 | GACG | 11 | – | 4 | – | – | ||
| DQ898064 | A... | 1 | – | 0 | – | – | ||
| DQ898065 | .G.. | 0 | – | 3 | – | – | ||
| DQ898066 | ..T. | 0 | – | 1 | – | – | ||
| DQ898067 | ...A | 0 | – | 1 | – | – | ||
| 1111222 | ||||||||
| 122239789 | 0.053⁎ | 0.061 | ||||||
| S. punctatus | 511385527 | |||||||
| DQ898068 | GTATTACTT | 20 | 2 | 12 | 64 | – | ||
| DQ898069 | .....G... | 1 | 0 | 0 | 0 | – | ||
| DQ898070 | ...C..... | 0 | 0 | 0 | 1 | – | ||
| DQ898071 | ......A.. | 0 | 0 | 0 | 1 | – | ||
| DQ898072 | .......C. | 0 | 0 | 0 | 1 | – | ||
| DQ898073 | A.......C | 0 | 0 | 0 | 1 | – | ||
| DQ898074 | .GT...... | 0 | 1 | 0 | 0 | – | ||
| DQ898092 | ....G.... | 0 | 0 | 0 | 1 | – | ||
| 11222222 | ||||||||
| 215233449 | 0.017 | −0.214 | ||||||
| S. spinus | 713713030 | |||||||
| DQ898079 | ACTTAATTC | 1 | – | – | 0 | 0 | ||
| DQ898080 | G..C....A | 0 | – | – | 0 | 1 | ||
| DQ898081 | G..CTGCCA | 0 | – | – | 0 | 1 | ||
| DQ898082 | GT....... | 0 | – | – | 0 | 1 | ||
| AY190545 | G.C...... | 0 | – | – | 1 | 2 | ||
| AY190554 | G........ | 0 | – | – | 1 | 0 | ||
| 122222 | ||||||||
| 25113699 | 0.105 | 0.316⁎ | ||||||
| S. vulpinus | 74767417 | |||||||
| DQ898084 | ACACAATT | 5 | – | 14 | 0 | – | ||
| DQ898085 | .....G.. | 3 | – | 4 | 0 | – | ||
| DQ898086 | ......C. | 1 | – | 0 | 0 | – | ||
| DQ898087 | G...G... | 0 | – | 1 | 0 | – | ||
| DQ898088 | .T...... | 0 | – | 1 | 0 | – | ||
| DQ898089 | .......C | 0 | – | 2 | 0 | – | ||
| AY190548 | .TGT.... | 0 | – | 0 | 15 | – |
⁎ ; **; ***.
1 Specimens from the Philippines identified as S. virgatus [10].
2 From the Philippines [10].
3 Specimen from the Philippines initially identified as S. vermiculatus (GenBank AY190556; [10]) but barcode-identified as S. lineatus (present results).
4 Includes one sequence of a Philippines specimen initially identified as S. punctatissimus (GenBank AY190543; [10]) but barcode-identified as S. punctatus (present results).
5 From the Philippines [10].
4 Discussion
Individual haplotypes of given nominal species generally formed tight clusters at the extremity of long branches, thus validating the current, morphologically-based taxonomy. While S. guttatus haplotypes appeared to cluster into a single clade, thus confirming its recognition as a separate species (in spite of apparent hybridization with S. lineatus (present results)), this group appeared to be nested within S. lineatus, suggesting that S. guttatus may be an offshoot of an ancestral proto-S. lineatus stock. The fact that the only haplotype sampled in the isolated S. lineatus population of the Maldives placed itself as outgroup to both S. guttatus and Indo-West Pacific S. lineatus, hints that the current taxon S. lineatus may include two geographically isolated populations with different evolutionary histories. Further insight into the genetic relationships of West-Pacific and Indian-Ocean populations of S. lineatus, requiring extensive sampling across the species' range is warranted, as this will inform us on the degree of genetic differentiation of Maldives S. lineatus vs. other S. lineatus populations. The two mitochondrial clades observed within S. fuscescens were separated from each other by an average nucleotide distance of the same order as that between S. guttatus and Indo-West Pacific S. lineatus, hinting at the possible occurrence of two distinct forms within the current taxon S. fuscescens in New Caledonia. To test that hypothesis requires further sampling of individuals, and the use of Mendelian markers to test for reproductive isolation.
Three pairs of presumed, sibling species with parapatric distribution were sampled for the present survey. Contrasted patterns of pairwise genetic differentiation were observed. The hypothesis of sibling species for the pair S. luridus/S. spinus was not supported. Letting aside the case of Maldives S. lineatus, the phylogenetic arrangement of S. guttatus/S. lineatus haplotypes was not inconsistent with the sibling-species hypothesis, but the fact that the two haplogroups did not appear as clearly separate clades also leaves open the possibility that the two species have not fully achieved reciprocal monophyly of their mitochondrial lineages. Nucleotide sequencing of a larger segment of the mitochondrial DNA may provide the necessary information to address that question. Last, there was no evidence of genetic differentiation of S. doliatus vs. S. virgatus. S. virgatus haplotypes were from individuals sampled in the Philippines, which border the zone of overlap with S. doliatus [3]. Since the two species are believed to hybridize [3,9], it is possible that those individuals identified as S. virgatus [10] harboured S. doliatus mitochondria as a result of introgression. In that case, further sampling of S. virgatus would be necessary, ideally outside the zone of overlap to sample ‘true’ S. virgatus mitochondria. Alternatively, the divergence between the two species may be recent, to the point that the sorting of their mitochondrial lineages is still not perceptible.
At the infra-specific scale, the degree of genetic differentiation was found to be lowest in S. argenteus and S. spinus, which of all Siganidae are the two species with the presumably longest pelagic, larval/prejuvenile stage [3] and also the widest distribution. Genetic-differentiation estimates in other species with more restricted ranges (S. doliatus, S. fuscescens, S. lineatus, S. puellus, S. punctatus, S. vulpinus) were all higher. Thus, distribution ranges, which primarily correlate with pelagic duration of larvae [3], also correlated well with the degree of genetic homogeneity among populations. These observations indirectly validate Woodland's (1990) hypothesis that dispersal is higher in species with longer larval stage in Siganidae [3].
The 300-bp cytochrome b gene fragment sequenced here proved a potential barcode for Siganidae, since the haplotypes sampled in a species generally clustered tightly, relative to the long intervals between species. In other terms, intraspecific pairwise nucleotide distances were distinctly lower than pairwise distances between haplotypes of different species, allowing efficient discrimination. This molecular marker allowed us to identify Siganus spp. larvae, all of which the current, morphology-based identification keys can only assign to family [26]. Most of the cytochrome b gene sequences that were available on GenBank in late 2005 when we started the present survey were from an unpublished study by D.G. Ochavillo and coauthors [10] whose objective was the identification and discrimination of juvenile Siganidae at the late pelagic presettlement stage. Our results allowed the barcode identification of two of the individuals sequenced by these authors, that is AY190543, initially identified as S. punctatissimus but which was found to be S. punctatus, and AY190556, which was identified as S. vermiculatus but here found to be S. lineatus. Barcoding of Siganidae will be useful in our investigations on the population and community ecology of shorefish larvae and juveniles, which require identification at the species level [11,27,28].
Acknowledgements
A part of this work was included in S. Lemer's M.Sci. memoir. Thanks to those who kindly provided samples: R.C. Anderson, F. Bonhomme, L. Carassou, J.-L. Justine, C. Mellin, G. Mou Tham, D. Lecchini, M. Leopold, J.E. Randall, P. Villard, and the Li-Khau family. Thanks to C. Rocher for technical help at the Dimar laboratory, and to P. Pruvost and L. Nandrin for curating voucher specimens at MNHN, Paris. Funded by the French government's ‘Fonds Pacifique pour le développement’, IRD-UR 128 ‘Biocomplexité des écosystèmes coralliens de l'Indo-Pacifique’, Dimar, and ZoNéCo-Opération ‘Jeunes poissons’.


