1 Introduction
Among all toxicants, cadmium (Cd) is considered one of the major pollutants in the current environment. It is a growing concern that the concentration of toxic heavy metals including cadmium is rapidly increasing in soil due to several manmade reasons such as mining, urban traffic, metal-working industries, mineral fertilizer, heating systems, waste incinerators and other sources [1,2]. Although Cd is highly toxic to plants, it can be taken up easily by roots and interrupt a wide range of biological processes, including enzyme activity [3,4], photosynthesis [5], respiration, transpiration [6], which can ultimately bring death.
Seed germination, a complex physiological process, is highly sensitive to metal pollution. The cadmium-induced inhibitory effect on seed germination has been well documented in many plant species, including rice [3,7–9]. To elucidate how plants defend themselves from the toxicity of cadmium and to investigate the cadmium-specific genes, extensive research has been conducted recently [10,11]. Even though numerous physiological and biochemical analyses have been conducted to investigate the responses of cadmium on different plant species [3,12–14], the proteomic approach has remained until now limited to the seed-germination stage. Identification of genes or proteins involved in biotic or abiotic stress responses is a fundamental step in understanding the molecular mechanisms of stress responses and developing transgenic plants with enhanced tolerance to stress. Two-dimensional polyacrylamide gel electrophoresis (2-DE) is an influential biotechnological tool for investigating differentially expressed protein profiles of seeds in response to abiotic stresses such as heavy metals [15,16] or to investigate germination-related proteins [17,18]. Proteomics, defined as the systematic analysis of the proteins expressed by the genome, is not only a powerful molecular tool for describing complete proteomes at the organelle, cell, organ, or tissue level, but also for comparing proteomes under different physiological conditions, such as exposure to heavy metal or other stressful environmental factors. Recently, few proteomic analyses have been conducted to investigate cadmium-regulated proteins in several plant species [19,20]. However, to the best of our knowledge, there has been no report on the cadmium-induced alternations of proteins at the germination stage of rice seeds.
In the present study, in order to gain a better understanding of the effect of cadmium on germination related proteins, 2-DE analysis was used as a powerful screening tool. Rice was selected as experimental material due to its well-organized database. Along with the proteomic analysis, other morphological and physiological analyses related to toxicity measurements were also conducted. The main goal of this study was to identify novel proteins that are highly upregulated at the germinating stage of rice seeds upon exposure to cadmium stress that can lead to a better understanding of the molecular basis of heavy-metal sensitivity in seeds at the period of germination.
2 Materials and methods
2.1 Plant material and germination assay
Mature rice seeds (Oryza sativa L. cv. Hwayeong) were collected from the Yeongnam Agriculture Research Institute, Milyang, Korea, and used as primary explants. Seeds were surface-sterilized as described previously [16]. Seed germination was tested on moist filter paper (Whatman No. 1). In each Petri dish (90-mm diameter), 30 seeds were randomly placed on the filter paper, then 10 ml of a cadmium solution, in a wide range of concentration levels (0.2 to 1.0 mM), were added. The controls were maintained by moistening the filter paper with 10 ml of deionized water. Cadmium chloride (CdCl2, Fluka) was used as cadmium salt and prepared freshly for the treatments. The seeds were germinated for four days under dark condition in a controlled (25 °C with 50% air humidity) growth chamber. In the present study, germination was considered when the coleoptiles were longer than 2 mm. Five replicates were carried out for each treatment, each one consisting of 30 bulked seeds.
2.2 Water content and biomass
The water content (WC) of the germinating seeds during treatment was determined using the following formula: [21]. Plant biomass was determined by weighing the seedlings, including the root and shoots. Prior to the fresh weight (FW) measurement, the seedlings were removed from the medium and washed with deionized water, followed by proper blotting between filter papers. After drying for 72 h at 60 °C in an incubator, the dry weight (DW) was measured.
2.3 Determination of cadmium content
To remove superficially metal, seedlings were washed five times with deionized water. The samples were then dried in an incubator at 60 °C for 72 h, weighed, and then ground to a fine powder. Accumulations of the cadmium in the samples were determined by ICP (inductively coupled plasma) emission spectrometry (Optima 3300XL, PerkinElmer, USA). Approximately 1 g of fine powder was used for each treatment, and the process was replicated three times.
2.4 Measurement of TBARS content
The level of lipid peroxidation in the germinating seeds was measured as the amount of thiobarbituric acid reactive substance (TBARS), determined by the thiobarbituric acid (TBA) reaction, according to Health and Packer [22]. In order to determine the TBARS concentrations from the treated and non-treated samples, the experimental procedure was followed as described previously [16].
2.5 Protein extraction
Proteins were extracted from the control (4-day-old seedlings) and treated seeds, including newly formed shoots after four days of germination. The roots were not included as a sample due to their absence in all treated samples. The polyethylene glycol (PEG)-mediated fractionated method [23] was used to remove abundant proteins from the samples. This method is currently widely used as a potential protein extraction method for rice samples for 2-DE analysis [16,24]. Proteins were extracted from the samples by a Mg/NP-40 buffer containing 0.5 M of Tris-HCl (pH 8.3), 2% v/v NP-40, 20 mM of MgCl2, 2% v/v β-mercaptoethanol, 1 mM of phenyl methyl sulfonyl fluoride and 1% w/v polyvinyl polypyrrolidone, followed by centrifugation at for 15 min. The proteins in the supernatant were subjected to PEG fractionation by addition of a 50% w/v PEG stock solution. The extract was adjusted to a final concentration of 15% w/v PEG. The mixed sample was incubated on ice for 30 min and then centrifuged at for 15 min. The supernatant was recovered and precipitated with acetone and stored in 1 ml aliquots at −20 °C until use. The acetone-precipitated sample was used for one-dimensional SDS-PAGE and 2-DE analyses. Protein content was measured according to Lowry et al. [25].
2.6 One-dimensional SDS-PAGE and 2-DE analysis
Equal amounts (15 μg) of proteins were separated on 12% polyacrylamide gels (5% staking and 12% separation gel), at a constant current of 20 mA. Before loading, the samples were heated in a boiling water bath for 30 s [16]. Separated polypeptides on the gel were visualized by staining with Coomassie Brilliant Blue (CBB), as described previously [16]. In order to achieve the precise sizing of the separated polypeptides, a protein molecular weight marker (SM 0431, Fermentas Inc., USA) was used.
2-DE (IEF in the first dimension, and SDS-PAGE in the second dimension) was performed as described previously [16]. The IEF gel mixture consisted of a 4.5% w/v acrylamide solution, 9.5 M of urea, 2% v/v NP-40, and 2.5% v/v pharmalytes (pH 3–10:pH 5–8:pH ; Amersham BioSciences, San Francisco, CA, USA). Each sample (350 μg of protein) was mixed with the sample buffer and loaded on an IEF gel (18-cm tube gel). SDS-PAGE in the second dimension was carried out as described by Laemmli [26]. The 2-DE gels were stained CBB, as described previously [16].
2.7 Image and data analysis
CBB-stained gels were used for image and data analysis. The intensities of the protein spots from waterlogged-treated samples were recorded as digitalized images using a high-resolution scanner (GS-800 Calibrated Imaging Densitometer; Bio-Rad). To control for subtle differences in sample loading, gel staining, and destining, the volume of each spot (i.e., spot abundance) was normalized to generate a relative volume, which was defined as the spot volume divided by the total volume of the entire set of gel spots [24]. Spots were detected and quantified with Bio-Rad PDQuest software (version 7.2; Bio-Rad, Hercules, CA, USA). After automated detection and matching, manual editing was carried out. A minimum of three gels was generated for each sample. Only spots that exhibited significant and reproducible changes were considered differentially expressed proteins.
2.8 In-gel digestion, protein identification and database search
Protein spots were excised from the CBB-stained gel and washed with 50% v/v ACN in 0.1 M of NH4HCO3, and vacuum dried. The gel fragments were reduced for 45 min at 55 °C in 10 mM of DTT in 0.1 M of NH4HCO3. After cooling, the DTT solution was immediately replaced with 55 mM of iodoacetamide in 0.1 M of NH4HCO3. After washing with 50% of ACN in 0.1 M of NH4HCO3, the dried gel pieces were swollen in a minimum 10-μL volume of digestion buffer, containing 25 mM of NH4HCO3 and 12.5 ng/μL trypsin (sequencing grade; Promega, WI, USA), and incubated overnight at 37 °C. Tryptic-digested peptides were extracted according to the protocol described by Lee et al. [24].
All peptide samples were analyzed using a Voyager-DE STR MALDI-TOF mass spectrometer (PerSeptive Biosystems, Framingham, MA, USA). The parameters for MALDI-TOF analysis were followed as described previously [27]. Briefly, parent ion masses were measured in the reflectron/delayed extraction mode, with an accelerating voltage of 20 kV, a grid voltage of 76.000%, a guide wire voltage of 0.01%, and a delay time of 150 ns. A two-point internal standard for calibration was used with des-Arg1-Bradykinin and neurotensin . Peptides were selected in the mass range of 500–3000 daltons (Da). For data processing, we used the PerSeptive-Grams software package. Database searches were performed using Protein Prospector (http://prospector.ucsf.edu). The following search parameters were applied using the National Center for Biotechnology Information (NCBI) protein sequence database: a mass tolerance of 50 ppm and one incomplete cleavage were allowed; acetylation of the N-terminus, alkylation of cysteine by carbamidomethylation, oxidation of methionine, and pyroGlu formation of N-terminal Gln residues were considered as possible modifications.
2.9 Statistical analysis
Physiological parameters and spot intensity data were statistically analyzed using Duncan's multiple range test or the Student's t-test. All results are presented as means ± standard error (SE) of at least three replicates. A P value of less than 0.05 was considered statistically significant.
3 Results and discussion
3.1 Toxic effects of cadmium on germination
In this study, we aimed to investigate the morphological, physiological, and proteomic changes in germinating rice seeds under cadmium stress. Therefore, as a preliminary experiment, germination responses and the relative biomass to various concentrations of cadmium were determined. Germination assay has been regarded as a basic experiment that can evaluate the toxicity of any kind of metal or chemical on plants [3,9,15,16,28–30]. The shoot growth of the germinating seeds gradually reduced with the increase of cadmium treatments (Fig. 1A). In addition, no root formation, however, was observed starting from 0.2 mM. In support with our observation, it has also been reported that lower concentration of cadmium inhibited the root growth more than it did on coleoptile growth of barely seeds prior to germination [31]. Our result also showed that the germination rate of the rice seeds responded differentially to cadmium toxicity (Fig. 1B) and was extremely reduced upon treatment with 0.4 to 1.0 mM of cadmium. Hence, we further determined the biomass production of the germinating rice seeds at the concentrations of 0.2 and 0.8 mM of cadmium (Fig. 2A). The results, presented in Fig. 2A, showed a significant variation regarding biomass production between the two treatments, as compared with the controls. According to these results, it could be concluded that cadmium has an adverse effect on rice-seed germination and that 1.0 mM of cadmium is the endpoint for the seed germination of the present rice cultivar. Genotypic difference of rice seedlings exposure to cadmium stress has been extensively studied, which allowed us to evidence that germination rate and subsequent responses of seedlings of different rice cultivars varied according to the level of cadmium treatment, and severely depressed under higher cadmium concentrations (0.5–2.0 mM) [9].

The effects of cadmium stress on rice seedlings. (A) Morphological responses of germinating rice seeds exposed to different concentration of cadmium. (B) Germination rate (%) and shoot length (cm) of young rice-seedlings subjected to four days of cadmium treatment. Data represents the averages ± SE of three independent experiments. Letters above the bars indicate a statistically significant difference (P<0.05) according to Duncan's multiple range test.

Cadmium induced physiological changes on germinating rise seeds. Water content (A), accumulations of cadmium in germinating rice seeds and their effect on biomass (B) and TBARS concentrations (C) were measured four days after germination. Data represents the averages ± SE of three independent experiments. Letters above the bars indicate a statistically significant difference (P < 0.05) according to Duncan's multiple range test.
3.2 Cadmium uptake and water status in germinating rice seedlings
The accumulations of cadmium in germinating rice seeds were determined after four days of treatment at cadmium levels of 0.2 and 0.8 mM. The data showed that cadmium uptake by germinating rice seeds was a linear function of cadmium concentrations in the solution (Fig. 2A). It has been shown that the metal uptake in plants is correlated with the increasing amount of metal in the growing medium or soil [9,15,16]. Water imbibition is the major factor for seed germination. Many factors, however, play a crucial role in inhibiting water uptake in seeds. In order to elucidate if cadmium generates an osmotic effect on germinating seeds, water content was measured. The water content in the germinating seeds decreased gradually and significantly according to the cadmium treatments, as compared to the control (Fig. 2B). To examine the osmotic effect of abiotic stress-treated plant tissues, the water content or relative water content was frequently measured [21], and it was observed that plant water status was highly affected by heavy-metal stress [6,15,16]. These results indicate that an excess level of cadmium has a toxic and an osmotic effect on seed germination.
3.3 Lipid peroxidation
Lipid peroxidation can be determined by the formation of TBARS as the decomposition product of polyunsaturated fatty acids (PUFAs), which are the main components of membrane lipids. Active oxygen species (AOS) can generate in vivo in plant cells due to a various types of abiotic stress, including heavy metals [32]. TBARS formation in plants, which are exposed to adverse environmental conditions, is a reliable indicator of cellular free-radical generation. We therefore measured the cellular TBARS concentrations as an indicator of cadmium-induced ROS formation. A greater accumulation of TBARS was observed under cadmium-treated samples that were statistically significant, as compared to the control (Fig. 2C). In addition, TBARS concentration was gradually increased in accordance with the concentration of cadmium in the medium. Significant increases of TBARS concentrations suggest that reactive oxygen species (ROS) may be generated by cadmium stress, which can initiate the peroxidation and destruction of lipids with subsequent damage [12,13].
3.4 SDS-PAGE analysis of cadmium-regulated proteins
As a preliminary experiment, cadmium-induced changes in protein patterns at germinating rice seeds were visualized by SDS-PAGE. The CBB-stained, one-dimensional protein pattern shown in Fig. 3 reveals the first differences between the control and treatments. The proteins patterns, between 116 and 45 kDa, and 25 to 14 kDa, which would be the major and metal binding proteins in rice seed, respectively, performed variations with respect to the control and treatments. According to our results, it was also observed that an increase in the concentrations of metal brought about drastic changes in the polypeptide patterns of rice seed. These results are in a good agreement with the previous report on maize [15] or in rice [16], where an increase in the amount of K2Cr2O7 or copper showed variations of seed proteome, respectively. SDS-PAGE is frequently used to visualize preliminary changes of polypeptide patterns by different types of heavy-metal-treated samples [15,16,33]. It is not sufficient to determine the pinpoint expanded changes in polypeptide patterns only by SDS-PAGE analysis. Therefore, 2-DE analysis was conducted as the next step of this experiment, in order to determine changes in detail of the protein patterns under cadmium stress.
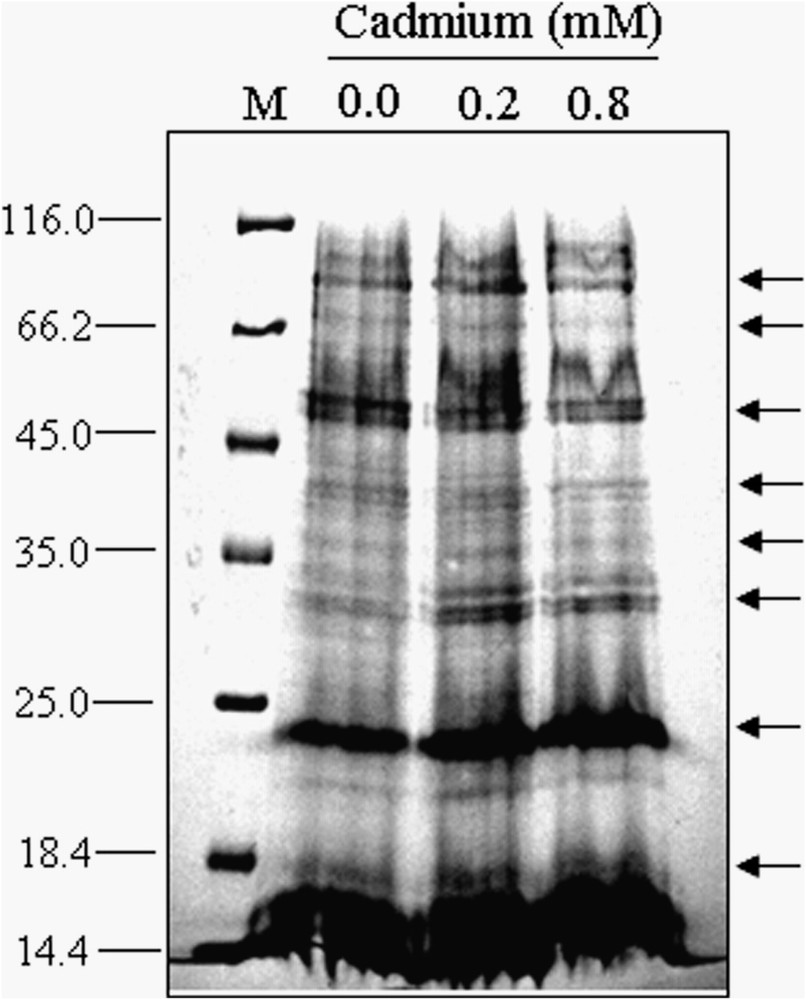
Changes in polypeptide patterns in germinating rice seeds after treatments with different concentrations of cadmium. Fifteen micrograms of proteins were loaded in each lane. Arrows on the right-hand side indicate some change of the polypeptide bands. Lane 1, control; lane 2, 0.2 mM and lane 3, 0.8 mM of cadmium-treated samples. The gel was stained with CBB.
3.5 2-DE analysis of cadmium-responsive proteins in germinating rice seedlings
2-DE analysis is regarded as a promising biotechnological research tool that can be successfully employed to analyze the regulation of seed proteins in response to several types of abiotic stresses [15,16,34] to investigate germination-related proteins [17,18] or to determine specific proteins [35]. Our SDS-PAGE experiment showed that a higher concentration of cadmium (0.8 mM) drastically changes the protein levels in germinating seeds; therefore, 0.8 mM of cadmium-treated samples were selected for 2-DE analysis. Glutelin is one of the most abundant housekeeping proteins in rice seed. The presence of abundance proteins in sample tissue sometimes creates problem in isolating differentially expressed rare proteins. A new pre-fractionation technique, mediated by polyethylene glycol (PEG), has been suggested [23,24] in order to overcome these kinds of difficulties. We therefore used the PEG supernatant fraction to obtain rare proteins by avoiding abundant proteins like glutelin. By using an equal volume of a fractionated protein sample from equal grams of fresh weight, 2-DE was carried out as described in Section 2. A high resolve of 2-DE gel pattern in the pI range between 4 and 7 was detected by CBB staining. More than 50 spots, however, exhibited variations between the control (4-day-old seedlings) and treatments (0.8 mM of cadmium) in CBB-stained gel. Among these, a total of 27 protein spots performed reproducible variations between control and treatment, and these proteins were subjected to MALDI-TOF analysis. Using PDQuest software (version 7.2; Bio-Rad, Hercules, CA, USA), a computer-aided image analysis program, the protein expression patterns and intensity of the protein spots between the control and treated samples were compared, and the differences are indicated in Fig. 4. Selected spots were subjected to MALDI-TOF mass spectrometry for peptide mass mapping. Six out of these spots did not match, while 21 upregulated proteins were identified, and the database search results are listed in Table 1. The representative 2-D gels in Fig. 5 show the distinct changes of all identified spots. Some of the identified proteins had different pI and Mr values than the theoretical ones. However, this phenomenon is commonly observed in proteomic analyses, and is likely a consequence of post-translational modifications, proteolytic cleavage, or matching proteins from different species [18,27,36].
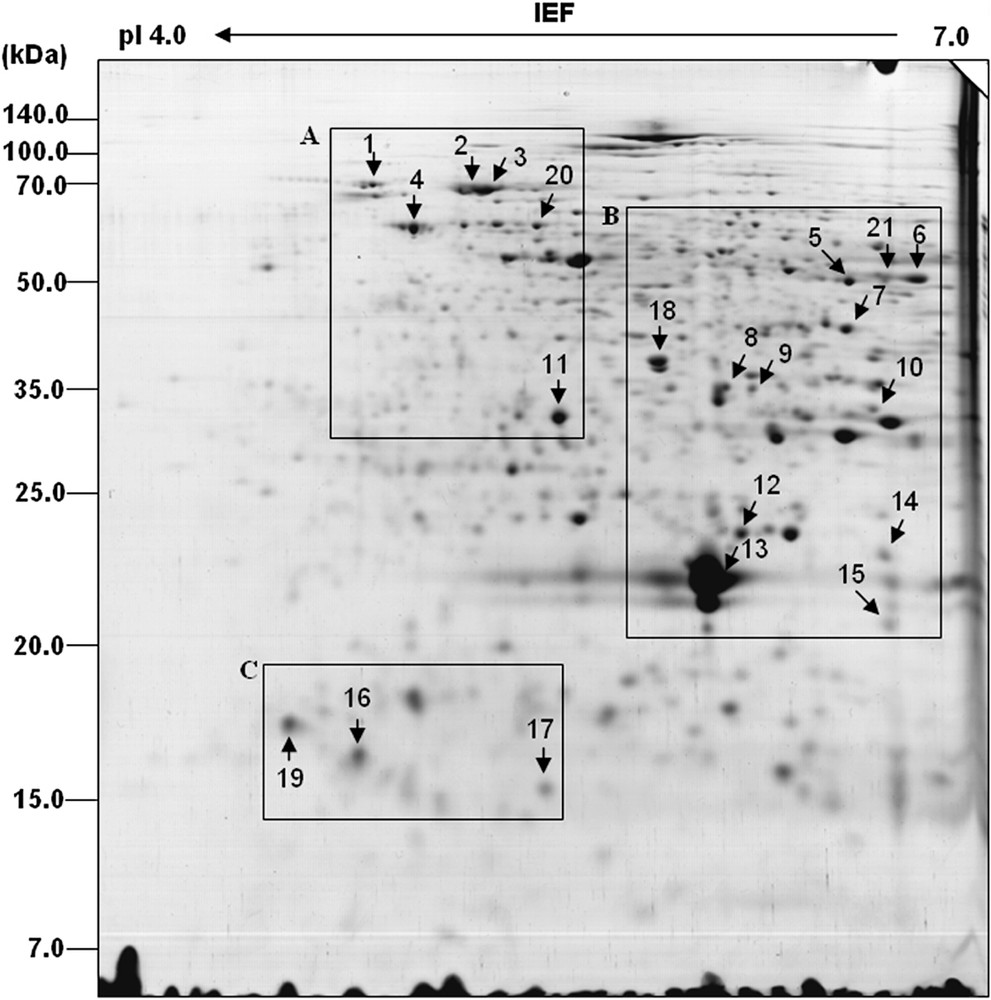
Rice-seed proteome patterns during the germination stage. A total of 350 μg of protein was extracted from the cadmium-treated four-day-old rice seedlings and separated by 2-DE, as described in Section 2, and visualized with CBB staining. Arrows indicate that the proteins were upregulated upon exposure to cadmium treatment. Cadmium-responsive proteins were located in the boxed areas (A–C).
Cadmium-induced upregulated proteins in germinating rice seeds identified by MALDI-TOF analysis
| Spot ID | Identified protein | Organism | Accession No.a | Theoretical/Observed | Mb | SC (%)c | MOWSE score | Fold changed | |
| pI | Mr (kDa) | ||||||||
| 1 | Putative receptor-like protein kinase | Oryza sativa | BAD32906 | 8.7/5.0 | 81/80 | 4 | 6 | 43.0 | 1.9 |
| 2 | Putative dnaK-type molecular chaperone BiP | Oryza sativa | XP_463871 | 5.1/5.3 | 73/73 | 11 | 22 | 1.208e+06 | 1.6 |
| 3 | Pentatricopeptide (PPR) repeat-containing-like protein | Oryza sativa | BAD72438 | 8.4/5.4 | 73/73 | 4 | 10 | 345 | 2.9 |
| 4 | Putative disulfide isomarase | Oryza sativa | BAA92322 | 4.8/5.2 | 33/57 | 11 | 38 | 9.268e+05 | 1.5 |
| 5 | Globulin-like protein | Oryza sativa | AAM33459 | 6.8/6.6 | 52/52 | 10 | 25 | 2.237e+04 | 1.7 |
| 6 | Globulin-like protein | Oryza sativa | AAM33459 | 6.8/6.8 | 52/45 | 4 | 12 | 454 | 2.8 |
| 7 | Myosin heavy chain-like | Oryza sativa | BAD68360 | 6.4/6.6 | 52/45 | 5 | 15 | 707 | 1.4 |
| 8 | Pentatricopeptide (PPR) repeat-containing-like protein | Oryza sativa | BAD27693 | 8.7/6.2 | 72/35 | 4 | 9 | 126 | 1.9 |
| 9 | Putative far-red impaired response protein | Oryza sativa | AAL58182 | 7.1/6.3 | 63/35 | 4 | 8 | 100.1 | 3.3 |
| 10 | Putative aldose reductase | Oryza sativa | AAU44244 | 6.3/6.7 | 35/32 | 4 | 20 | 884 | 1.9 |
| 11 | Glyoxalase I | Oryza sativa | XP_480480 | 5.5/5.7 | 32/32 | 10 | 45 | 4556 | 3.2 |
| 12 | Peroxiredoxin | Oryza sativa | AAQ01200 | 6.0/6.3 | 24/23 | 9 | 39 | 1.791e+04 | 1.7 |
| 13 | 19 kDa globulin precursor | Oryza sativa | CAA45400 | 7.5/6.7 | 20/23 | 5 | 25 | 268 | 1.8 |
| 14 | 19 kDa globulin precursor | Oryza sativa | CAA45400 | 7.5/6.7 | 21/23.5 | 6 | 25 | 1085 | 3.7 |
| 15 | Putative retroelement | Oryza sativa | NP_920503 | 4.9/6.7 | 25/21 | 4 | 17 | 345 | 1.6 |
| 16 | Ulp1 protease-like | Oryza sativa | BAD35625 | 6.4/4.9 | 19/17 | 5 | 21 | 455 | 21.0 |
| 17 | Hypothetical protein | Oryza sativa | XP_465897 | 7.2/5.7 | 20/16 | 4 | 31 | 218 | 2.3 |
| 18 | OSJNBa0055H05.9 | Oryza sativa | CAE02722 | 6.5/6.0 | 51/42 | 4 | 11 | 509 | 2.2 |
| 19 | Chaperone protein HchA | Staphylococcus aureus | NP_371075 | 4.9/4.7 | 32/18 | 4 | 18 | 30.9 | 11.0 |
| 20 | ATP synthase subunit B | Bdellovibrio bacteriovorus | NP_970598 | 5.3/5.6 | 50/57 | 4 | 16 | 20.6 | 2.7 |
| 21 | NPR2 | Saccharomyces cerevisiae | CAA55721 | 8.9/6.75 | 70/52 | 5 | 15 | 27.5 | 1.9 |
a NCBI accession No.
b Number of peptide matches.
c Sequence coverage.
d Protein fold increased compared to the control.
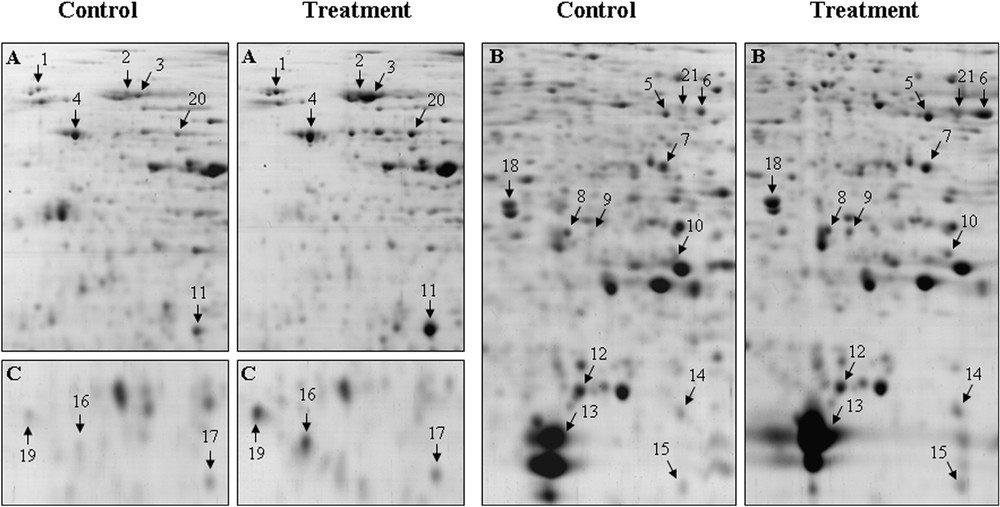
Magnified views of the upregulated proteins in rice seeds in response to cadmium stress. A–C indicate the boxed areas marked in Fig. 4. Control, four-day-old rice seedlings and treatment, four-day-old rice seedlings exposed to 0.8 mM of cadmium. Arrows indicate temporal changes of the differentially expressed proteins after cadmium stress.
3.6 Cadmium-induced upregulated proteins
Plants have evolved a complex sensory mechanism in order to monitor their surroundings and to adapt their growth and development to the prevailing environmental conditions [37]. Heavy metals have an adverse effect on normal plant growth and development, and regulate a wide range of genes that are involved in many cellular responses [38]. The binding of heavy metals to sulphydryl groups in proteins, which leads to an inhibition of an activity or a disruption of a structure, or the displacement of an essential element, or the production of free radicals and reactive oxygen species are the possible ways for heavy metals to disrupt normal physiological and biochemical functions of cells [39]. Therefore, we identified a group of upregulated proteins that are involved in RNA processing, signal transduction, protein biosynthesis and translation, antioxidant, defense and detoxification or/and germination processes.
A group of regulatory proteins, including receptor-like protein kinase (RLK), DnaK-type molecular chaperon Bip, pentatricopeptide (PPR) repeat-containing-like proteins, putative far-red impaired response protein, putative retroelement and Ulp1 protease-like proteins, are markedly upregulated or newly synthesized by cadmium stress. The RLK genes constitute a very large family in the plant genome and play various roles in plant growth and development, and defense reactions such as meristem control, phytohormone signaling, disease resistance, nodulation and self-recognition [40]. Even though this protein was also observed in the control sample, its upregulation by cadmium treatment suggests a possible correlation with heavy-metal toxicity. Consistently with our results, it has been reported [41] that the overexpression of a cell-wall-associated receptor kinase in Arabidopsis enhanced metal toxicity. Molecular chaperons such as Hsp70 chaperons are expressed in response to a wide range of abiotic stresses [42] and they play a crucial role against stress, by re-establishing normal protein confirmation and thus, cellular homeostasis [43]. We identified a putative DnaK-type molecular chaperon Bip (spot 2), which increased 1.6 fold in response to cadmium stress. The DnaK Bip protein is a subfamily of Hsp70 that could prevent proteins from unfolding, mainly in endoplasmic reticulum under stress conditions [42]. It has also been reported that Bip proteins might directly alleviate cells from chemical stress [44]. We identified two putative pentatricopeptide (PPR) repeat-containing-like proteins (spot 3 and 8), from both the control and treated samples. Even though the identity of these two spots was the same, their molecular mass and pI were a bit different. In 2-DE analysis, this phenomenon has been frequently observed due to post-translational modification or proteolytic cleavage [18]. PPR proteins represent a large family and the vast majority of these proteins are predicted to be targeted by either mitochondria or chloroplasts [45]. Yan et al. [36] observed that the PPR protein has also been induced under abiotic stress in rice leaves, thus suggesting their participation in RNA processing under stress conditions. In our investigation, we identified a putative far-red impaired response protein (spot 9) that showed a higher intensity (3.5-fold) compared to the control. The relation between the protein and metal stress, however, is completely unknown. A putative retroelement protein (spot 15) was identified from both the control and cadmium-treated samples. Retroelements are frequently observed as ubiquitous mobile genetic entities in eukaryotic genomes. In the rice genome sequence, a total of 217 full-length retroelement sequences were identified [46]. The retro-transposon might play an important role in the re-organization of genes and the optimization of gene expression in response to various environmental stimuli [47]. There is evidence to suggest that biotic and abiotic stresses are related to an increment of their activity [48]. Several cellular physiological processes, including selective protein degradation, DNA repair, cell-cycle control, apoptosis and response to biotic and abiotic stress [49] have been implicated in plants by ubiquitin (Ub) and ubiquitin-like (Ubl) protein pathways. An Ulp1 protease-like protein (spot 16) has been identified from both control and treated samples. Metal or oxidative stresses that induced the upregulation of the ubiquitin-like protein have also been reported in tomato plants [16,49]. These results clearly indicate that the Ulp1 protease-like protein is usually related with the differentiation and cell-cycle regulation during seed germination and that their upregulation was caused by metal-inductive oxidative stress.
We identified a number of stress and detoxification-related proteins that were markedly upregulated in response to cadmium stress, consisting of species such as putative disulfide isomarase (spot 4), myosin heavy chain-like protein (spot 7), putative aldose reductase (spot 10), glyoxalase I (spots 11), and peroxiredoxin (spot 12).
The protein disulfide isomerase (PDI) is a ubiquitous, multi-functional enzyme that is not only present in the lumen of the endoplasmic reticulum (ER), but it is also co-localized with seed storage proteins in dense protein bodies [50]. We detected a PDI (spot 4) from both the control and treated samples. In Arabidopsis seeds' proteomics analysis, PDI has been identified as a constant polypeptide whose abundance was not changed prior to germination [17]. However, in our investigation PDI was identified as an upregulated protein in response to cadmium. This result suggests that PDI may play a specific role for protecting the cells under cadmium stress. In support to this hypothesis, it has been shown that the PDI forms a stable dimer in the presence of physiological levels of Zn and other thiophilic divalents such as Cd, suggesting that its interaction with divalent ions could play a role in the regulation of effective concentrations of these metal ions, thus protecting the plant against metal toxicity [51]. A myosin heavy chain-like protein (spot 7) was detected from both control and treated samples. In the present study, we are unable to clarify the real function of this protein in response to cadmium treatment; however, we assumed that, as a cytoskeletal protein, it might be related to the cell cycle and/or morphogenesis. Therefore, it could be observed from both control and treated samples. Cytoskeleton and its modulators are known to play a key role in cell morphogenesis [52]. The cytoskeleton is, in part, restructured as a result of the phosphorylation of the three threonine residues in the tail region of the myosin II heavy chain, which regulates its state of assembly [53]. Exposure to heavy metals also induces change in the tyrosine phosphorylation of actin and in actin cytoskeletal organization [54]. Recently, this protein has also been identified from chilled-stress-treated rice leaves [36], which suggest that it may play a role against oxidative stress. Aldose reductase is the key enzyme in the polyol pathway, leading from glucose to sorbitol, which is exclusively expressed in embryos and is regulated by ABA and GA [55]. It has also been reported that aldose reductase is a common osmolyte that helps to balance cell osmotic strength with that of abiotic stress [55]. Aldose reductase has also been identified in Arabidopsis seed proteomic analysis as an unchanged protein during germination [17]; however, in the present study, it was upregulated nearly two times under cadmium stress compared to the control. Oberschall et al. [56] observed that a wide range of stress factors, including cadmium, could trigger the synthesis of the alfalfa aldose reductase. In support of this finding, Hegedus et al. [57] reported that alfalfa aldose reductase overexpressing tobacco plants showed an enhanced tolerance to cadmium stress. These results suggest that aldose reductase could be used as an alternative biomarker for cadmium stress. Spot 11 was identified as glyoxalase I, which is the key enzyme of the glyoxalase pathway. The role of glyoxalase I is well documented in the detoxification of methylglyoxal, which is formed due to several types of abiotic stress [58]. Moreover, enhanced cadmium-tolerant tobacco plants were obtained by the overexpression of the glyoxalase pathway-related genes [59]. In maize or rice seed proteomic analysis, the upregulation of the glyoxalase I was also observed when seeds were exposed to high concentrations of potassium dichromate [15] or copper [16], respectively. In addition, glyoxalase was identified as a constant protein in Arabidopsis seed protein analysis prior to germination and priming [17]. Taken together, these results suggest that glyoxalase I plays a crucial role in metal toxicity detoxification. Peroxiredoxins (Prxs), which are abundant low-efficiency peroxidases, have a diverse range of cellular functions such as an antioxidant and an endogenous regulator of apoptosis [60], antioxidant [61], and a molecular chaperon [62]. We detected the peroxiredoxin protein (spot 12) from both control and cadmium-treated samples. Recently, it has been observed that the peroxiredoxin-type protein was also upregulated in maize roots [63], seeds [15], and rice seeds [16], in response to arsenic, potassium dichromate, and copper, respectively.
A group of major seed-storage proteins, such as globulins (spots 5, 6) and their precursors (spots 13, 14), were identified from both control and treated samples. The presence of globulin and their precursors has been frequently observed in rice and other cereals seed proteome analyses [15,35]. In germinating maize seed, the upregulation of the globulin precursors was also detected by potassium dichromate treatment [15], which suggests the activation of storage proteins.
Two proteins (spots 17 and 18) were identified as having unknown function, and additional studies are needed to address their putative role in the response to cadmium stress. In addition, the PMF data of three proteins (spots 19–21) were not matched with the rice, but showed considerable sequence coverage with the other microorganisms. Interestingly, the putative functions of these proteins are also stress-related. However, at present, their function in response to cadmium-stress response in rice seeds is unclear.
4 Conclusions
In this study, we have pointed out that the inhibition of seed germination upon exposure to cadmium not only interfered with water uptake by seeds, but was also disrupted by oxidative stress, which may be interrupted other physiological processes. Physiological data showed a good correlation with the proteomic analysis. A total of 21 proteins were identified that were differentially expressed in response to cadmium stress. A greater accumulation of antioxidant and of other stress-related regulatory proteins in the germinating rice seeds upon exposure to cadmium stress suggests that they might work corporately to establish a new homeostasis in response to cadmium, as well as to other types of heavy-metal stress. While the present study is a good starting point for understanding the molecular response of seeds to metal stress, further investigation and advanced functional analysis of the differentially regulated proteins identified in this study are required to determine their specific role in rice seeds' response to cadmium stress. An analysis of sequential cadmium-stress treatment of sensitive and tolerant rice cultivars would also lead to a better overall understanding of the responses of rice seedlings to metal stresses.
Acknowledgements
Prof. Pil Joo Kim from the Division of Applied Life Sciences (BK21), Gyeongsang National University, Jinju 660–701, Korea, is gratefully acknowledged for the analysis of cadmium content. Dr. Nagib Ahsan is supported by a post-doctoral grant from the BK21 program, and S.-H. Lee and D.-G. Lee, are supported by a scholarship from the BK21 program, Ministry of Education & Human Resources Development, Korea. This work is supported partially by a grant from the BioGreen 21 Program, Rural Development Administration, Korea.


