1 Introduction
Soil mineral nitrogen availability is generally heterogeneous and fluctuating which hampers sustained N acquisition by the plant root systems. On the one hand, mineral nitrogen present around the roots, arising from either added fertilizer or mineralized organic matter, can be depleted rapidly through leaching. On the other hand, plant roots experience frequently either abiotic stress (water and temperature stress) or pathogen attack that may have important impacts on root structures and their activities. Plants have developed adaptive responses modulating the efficiency of root N acquisition as a function of both external nitrogen availability and the nutritional status of the whole plant.
A general model of control of plant root acquisition involving local signaling and the systemic action of long distance signals of the plant N status has been proposed:
- – Nitrate locally regulates plant intake, metabolism and development by: (i) inducing the expression of proteins required for its utilization by the plant; and (ii) stimulating lateral root growth, root branching and thus soil exploration [1]. Transcriptomic approaches on Arabidopsis thaliana, tomato, and rice have allowed the identification of more than a thousand genes differentially expressed upon supply [2–7] and a robust network associated with the signal has been proposed [8].
- – Mechanisms involved in the long distance control of root acquisition by the N status of the plant are much less documented than those involved in the local regulation by . Data obtained on tobacco (Nicotiana plumbaginifolia) and Arabidopsis shown that down-regulation of transporters is exerted by downstream N metabolites of the whole plant [9–13]. Exogenous supply of amino acids strongly represses both uptake and the expression of key transporter genes in the roots [14–17]. A systemic signaling modulating lateral root development in response to changes in supply has also been proposed [18].
Plant N may be acquired in several forms not only mineral ( and ) or organic (amino acid and peptides) but also as N2 through symbiotic interaction with Rhizobium bacteria in legumes. The capacity to establish symbiotic N2 fixation has an important impact on the adaptation of legumes to low mineral nitrogen supply because N2 is an unlimited source which is continuously and uniformly present in the plant environment. However, because symbiotic nitrogen fixation requires high level of carbon allocation and is highly sensitive to local stresses generally found in soil, N2 fixation capacities of the leguminous roots are frequently limited.
How acquisitions of , and N2 in leguminous plant are coordinately regulated to fulfill the plant N demand is not known. The fact that the assimilatory pathways involved in the acquisition of these N sources share the same end-product might suggest that the three pathways of root N acquisition might be under a general control exerted by a systemic signaling pathway related to the level of downstream product of N assimilation in the whole plant [11,19,20]. Reports indicating that downstream N metabolite (glutamine or/and asparagine) concentrations either in roots or their medium are inversely correlated with both uptake and nodule N2 fixation activity [21–24] supports this hypothesis. Such a model would predict that the inhibitory effect of high mineral N on N2 fixation by suppressing nodulation and inhibiting N2 fixation activity might be related to a systemic feed back regulation of downstream metabolites [19,23]. Against this hypothesis of a common systemic regulation for all N sources, split root experiments shown that uptake and lateral root growth were not stimulated on the untreated side of roots in response to localized N-limitation on the other root part, indicating at least in this case that the release of feedback repression mechanism does not lead to a significant increase of acquisition which may compensate the local N limitation as it is the case in fed plants [10,18]. Furthermore, several reports in legume concluded that the inhibition of N2 acquisition by mineral N is mainly local, suggesting that nitrate itself, rather than a product of N assimilation, may act as an inhibitor [25]. In the supernodulating mutants of pea (Pisum sativum L.), soybean (Glycine max L. Merr), Lotus japonicus, Medicago truncatula (Mtr), mutated on the same orthologous gene named MtSUNN in Mtr and HAR1 in Lotus Japonicus [26,27], graft experiments demonstrated that the deficiency in autoregulation of nodulation is under shoot control. The fact that these mutants also display a nitrate tolerant nodulation feature suggests some common systemic control for autoregulation and nitrate tolerance [27–30]. Nevertheless, the regulatory mechanisms involved in these responses are poorly understood and still remain largely unknown at the molecular level.
In the particular case of legumes, understanding plant adaptation to N limitation necessitates also to take into account specifically the bacterial symbiotic partners and strategies that are used by the plant to adapt symbiotic organ structure and function to its N demand. Interestingly many steps of the signaling cascade involved in the initiation of the plant–Rhizobium symbiosis and nodule establishment have been characterized in the past 10 years [31–35]. In the hypernodulating sunn mutant of Mtr, a receptor like kinase involved in the systemic control of autoregulation of nodulation has been identified [27]. However, the role of this regulatory pathway in the control of legume N acquisition by the N status of the plant is far to be understood. Intriguingly, these mutants display a growth reduction of both shoots and roots which probably results from the competition for assimilates between these plant organs and the buildup and functioning of nodules, illustrating the necessity of a tight integration of N and C acquisition at the whole plant level.
Root architecture is a major determinant of the size of the root-soil interface and of the resulting water and nutrients acquisition by the plant. Several genes of Arabidopsis thaliana involved in root development through hormonal signals and/or nutrient perception were reviewed by Casson and Lindsey [36]. Much less is known concerning legume root development. A spontaneous genetic variability for root length and biomass has recently been described in pea [37–39]. Conversely, genetic variability of the symbiotic microbial partner has been shown to also influence the development of nodules, roots and shoots [40]. An induced branched root mutant and a long root mutant were also reported in pea [41]. Most of these studies identified interactions between development of roots, nodules and shoot suggesting some common regulations. Genes MtLAX1, MtCRE1, MtSICKLE, Mt SUNN and MtLATD of Mtr were shown to be involved in root and nodule development [42–46]. The fact that in the absence of Rhizobium, supernodulated mutants of Mtr and Lotus japonicus (sunn and har1 respectively) display a short root length phenotype, associated in the case of har1 to an enhanced number of lateral roots [26,27,30] revealed another common regulation pathway for root and nodule development.
Although numerous works allowed the identification of genes involved either in N acquisition or in the development of the structures involved in N acquisition (roots and nodules), the mechanisms behind the integration of their function at the plant level still remain obscure. Plant nitrogen nutrition is a highly integrated process which implies: (i) signalization between the plant organs (leaves and roots); (ii) interactions between developmental and functional process of these organs; (iii) interactions with plant N/C metabolism; and (iv) interactions between the various N acquisition pathways.
The present article aims at giving a brief synthesis of an integrative approach, gathering works conducted at different scales of investigation. The biological question is addressed through changes in the scale level from the gene to the plant level, using common genotypes, addressing the question of whole functioning and of plant/Rhizobium interactions. In order to unravel the control of N acquisition in legumes by the N status of the plant the approach was conducted with a particular effort to: (i) decipher the regulating elements that are either common or specific of the three main types of N nutrition (, and N2 fixation) and their shortage, and (ii) model the emerging properties occurring at the whole plant level. These mechanisms were investigated in the legume Mtr, a species able to acquire nitrogen as , or N2 (through symbiotic infection with Rhizobium). Mtr has been chosen as a model because of its evolutive proximity and syntheny with several cultivated legume crops, its small genome size, the ease of its transformation, the availability of mutant resources and the forthcoming genomic sequence [47,48].
2 Interaction between organs
An obvious level of investigation is the level of the organs specialized in the N acquisition (roots and nodules). Nevertheless, a possible control by the N demand of the whole organism relations between these organs and the other parts of the plant has to be considered. Experimentally, split root systems allow one: (1) to evaluate the effect on the whole plant of short term fluctuations of N availability applied on a localized part of the roots; and (2) to evaluate the systemic responses triggered at the whole plant level that are revealed on the untreated side. The responses of roots normally supplied with various nitrogen sources (1 mM , 1 mM , or air) to contrasted level of N supply on the other side have been investigated. Three types of N nutritional situations were compared referred as “Control”, “N-sufficient” and “N-limited”. In “Control plants” both sides of the split-root were normally supplied with 1 mM , 1 mM or air. In “N-sufficient” plants one side of the root system was supplied with a high level of nitrogen (10 mM NH4NO3) whereas in “N-limited” plants the N source was absent of this compartment (air was substituted by Argon/O2 in case of nodulated plant). Short term responses (4 days) affecting the acquisition machinery (, , uptake and symbiotic N2 fixation) were studied at first.
Nitrate, ammonium and N2acquisition are repressed by systemic signaling: The addition of high level of NH4NO3 to one side of the root system induced a strong repression of the acquisition capacity of the other side whatever the nature of the N source (, , air) at that side (Fig. 1). This indicates that: (i) the three pathways involved in N acquisition are under feedback repression mechanism(s); and (ii) these feed back mechanism(s) involve systemic signaling pathways that adjust the acquisition capacities of the roots to the N demand of the whole plant. Parts of the regulatory mechanisms may be common for the three acquisition pathways.
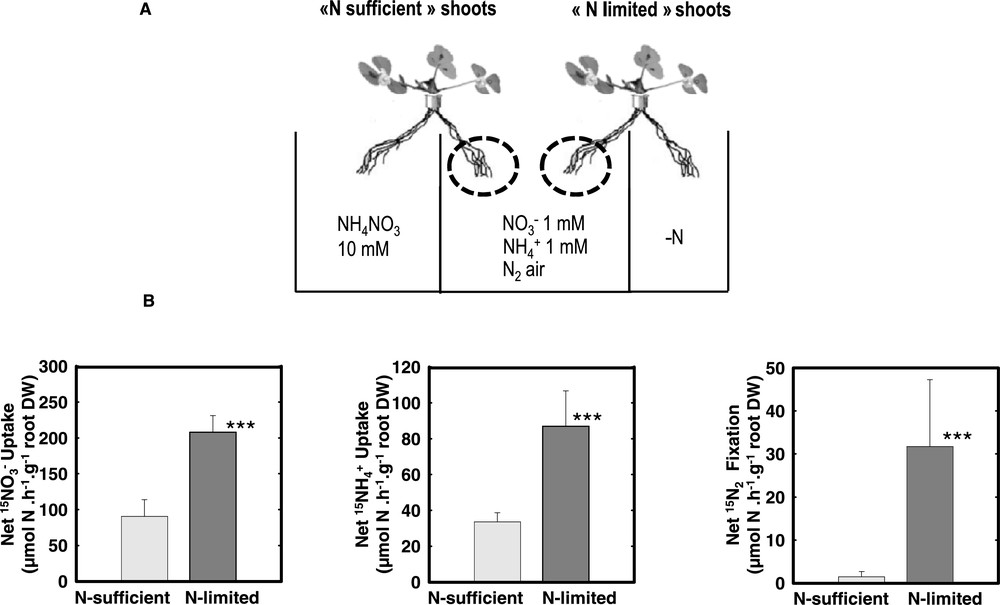
Nitrogen acquisition of the untreated roots of N-satisfied and N-limited plants. (A) Split-root systems were used to compare N-limited and N-satisfied hydroponically grown plants. Plants were fed with either 1 mM NO3, 1 mM NH4 or fixing N2 (nodulated in presence of Rhizobium meliloti). Over a period of 4 days, N-sufficient plants received a concentrated (10 mM) NH4NO3 solution on the treated-side of the root system while in N-limited plant the N source was removed from the treated-side of the root system (N-free solution for and plants, replacement of normal air by a 80%/20% Ar/O2 mixture in the root atmosphere for N2-fixing plants). (B) N acquisition of the untreated roots in the center of the split root measured using 15N labelling. The values are the means of six replicates of one biological repeat. They are representative of three independent biological repeats. Vertical bars indicate SD. ***Significant difference according t-test (P < 0.001). Adapted from Ruffel et al. [49].
Molecular responses associated with these physiological responses have been characterized by a transcriptomic approach: To further investigate the molecular basis of the short term control of root N acquisition to variations of N status, transcriptomes of roots and shoots of N-limited and N-sufficient plants supplied with , , or air were compared using affymetrix chips and real time Q-PCR platform. Gene lists of differentially expressed genes have been identified for the three sources [49] (Fig. 2). In roots, important groups of these genes are coding proteins related to the specific acquisition machinery of roots or nodules (transporters, assimilatory enzymes). Some are related to root or nodule development suggesting that both the function and the structures are under the control of systemic N signaling. Despite a similarity of the responses of the three pathways involved in N acquisition to these treatments at the physiological level, the transcriptomic study has revealed that at the molecular level genes networks involved in these responses in root and in shoots are rather specific of the N source present in the environment of the roots. Of course this does not rule out the possibility that some elements involved in the sensing of the plant N status might be common for the three N sources status and that these elements constitute potential key molecular components of the systemic signals regulating root N intake. All together these results may suggest that the plant response may be the result of the interaction of two signaling pathways (presumably systemic) related to the nature of the N source and to the level of downstream N metabolites respectively that may determine the response of plant roots and leaves to N availability modulation.
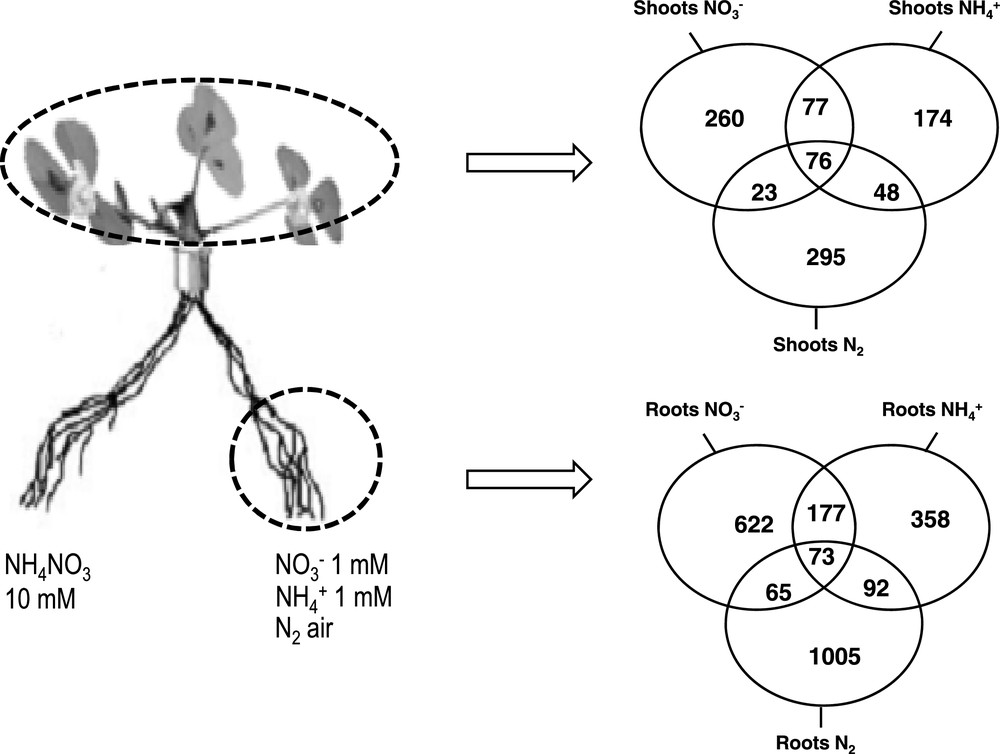
Venn diagrams of genes identified as differentially expressed by microarray experiments in shoots (A) and in roots (B) of the plants described in Fig. 1. Adapted from Ruffel et al. [47]. Data are based on the analysis of two independent experimental repeats. Affymetrix GeneChipMedicago Genome Array contains over 61,200 probe sets: 32,167 based on the EST/mRNA and chloroplast gene sequences of Mtr., 18,733 based on the partial genomic sequence of Mtr and 1896 based on the EST/mRNA sequence of Medicago sativa and 8305 based on the genomic sequence of S. meliloti (http://www.affymetrix.com). Total RNAs of the various organs were used for microarray experiments. CRNA synthesis, labeling, hybridization, washing, staining, and scanning arrays were carried out as recommended by the manufacturer's instruction manual (http://www.affymetrix.com). Normalization and data mining were described by Ruffel et al. [49].
When plants experience a local N limitation, onlyacquisition is able to display an efficient adaptive response that may efficiency compensate the N deficit: Upon a short (i.e. 4 days) limitation on one side of split root systems, nitrate fed plants maintain their overall N acquisition and growth. This compensation for a temporary N deficit is mediated by the stimulation of the uptake affinity and capacity of the other side of the root system still fed with (Fig. 3). This short term response is easily interpreted as the result of a release of the feed back repression exerted by N metabolites. However, despite the fact that such mechanism exists for the three types of N acquisition intriguingly only fed plants display this capacity to compensate a local N limitation. The consequence is important for the plant because only acquisition may allow the whole plant to maintain its growth under such conditions, whereas in , N2 fixing plants, local N limitation results rapidly in a decrease of the overall N acquisition of the plant and therefore limits plant growth. As and N2 fixation require important C metabolites flux to the roots to be assimilated, a logical cause might have been a limitation of compensatory response by carbon availability. However, sucrose feeding has ruled out this hypothesis, suggesting that the absence of response was presumably due to an overall limitation of the uptake and N2 fixation capacities of the plant roots that are not as efficient as in the case of .
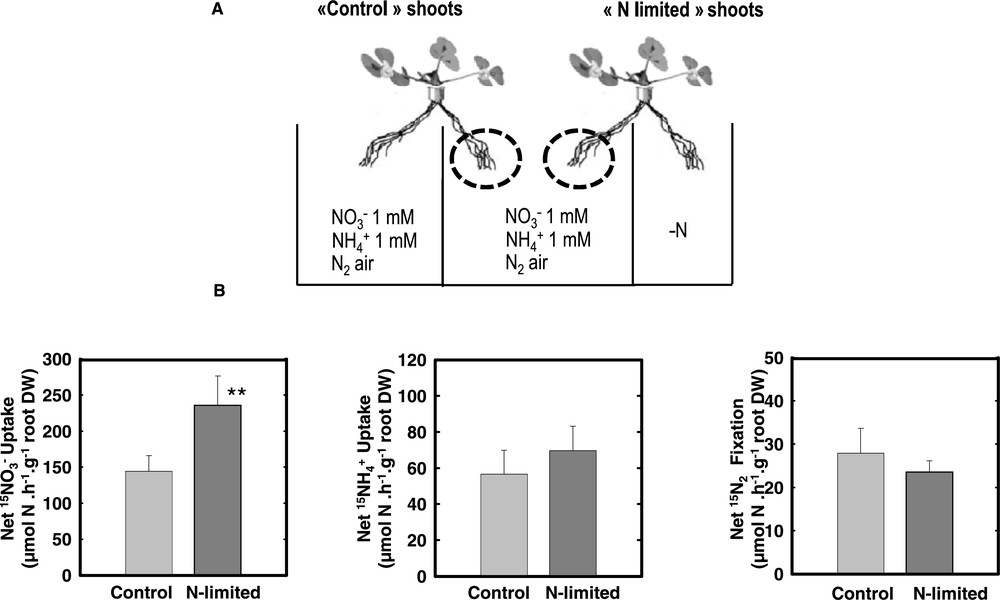
Nitrogen acquisition of the untreated roots of Control and N-limited plants. (A) Split-root systems were used to compare N-limited and N-satisfied hydroponically grown plants. Plants were fed either 1 mM NO3, 1 mM NH4 or fixing N2 (nodulated in presence of Rhizobium meliloti) in split-root systems similar to those in Fig. 1. While control plants received this N regime on both sides of their root system, the N source was removed to the treated-side of the roots of N-limited plants as in Fig. 1. C and L roots belong to Control or N-limited plants but were continuously exposed to the same local environment during the treatment. (B) N acquisition of the untreated roots in the center of the split root measured using 15N labelling. The values are the means of six replicates of one biological repeat. They are representative of three independent biological repeats. Vertical bars indicate SD. **Significant difference according t-test (P < 0.01). Adapted from Ruffel et al. [49].
Molecular responses identified by the transcriptomic approach are associated to the functional response of the organs: Representative subpopulations of root transcripts that have been identified in our transcriptomic studies (comparison between N-sufficient vs N-limited plant) were selected and studied in response to local N limitation (comparison between “Control” and “N-limited” plants). Interestingly, the differential expression of these genes is well correlated to the adaptive response observed in those plants: only in the case of fed plants a large proportion (>50%) of the genes identified as responding to systemic repression signals were found also differentially expressed in response to local N limitation. In contrast, in the case of or N2 fed plants that do not display functional response, most of the genes identified as regulated by the systemic signal (N-sufficient vs N-limited) comparison did not respond to local N limitation [49].
3 Integration of developmental and functional responses of plant organs
A second level of integration is to take into account developmental responses that may affect (both in time and space) structures involved in N acquisition (nodules and roots). Although response involving initiation of new structures may be initiated very rapidly after beginning of N shortage, to be efficient they generally require more time than response modulating the activity of pre-existing structure. Therefore N-limitation treatments were applied on fed and N2 fed plants in split root systems on a larger time scale than before (up to 16 days) in order to potentially reveal these responses. The long-distance effects of N limitation were studied by monitoring N intake fluxes (15N labeling), root and nodule morphometry, nodule initiation (by the use of pENOD11::GUS reporter gene), C allocation within the plant (13C labeling).
In contrast with nitrate fed plants, N2 fixing plants lack the ability to trigger a fast up-regulation of N2 fixation. Our results indicate that in both fed and N2 fed plants, N limitation triggers a systemic signaling which results in developmental response that may compensate the N deficit. In fed plants, roots of N-limited plants strongly proliferate in the nitrate containing compartment after 14 days (as compared to nitrate fed roots of control plants) to the expense of N-limited roots that stop growing. Together with the up regulation of the uptake capacities of these roots that was described above, this response leads to a very efficient compensation of N limitation, that allows the N-limited fed plant to maintain their growth at the same level as the control fed plants. Nevertheless, although less efficient as in the case of N fed plants, a long term response relying on long term plasticity of nodule development can be revealed in N2 fed plants exposed to local N limitation. On the untreated roots, this response involved firstly the expansion of pre-existing nodule and secondly the initiation of new nodules demonstrating that both late and early stages of nodule development are targeted by systemic signals related to the N status of the plant. Interestingly the use of the Mt sunn hypernodulating mutant indicated that both responses may be mediated by distinct pathways, the first one being probably sunn independent whereas the second one sunn dependent. Although the mutant displays a higher number of nodules, they have a lower level of fixation activity (overall N2 fixation capacity of the plant being at the level of the wild type). Surprisingly, unlike the wild type, when the mutant is exposed to N limitation it is able to up-regulate rapidly nodule N2 fixation specific activity and compensate partially the N deficit suggesting that the presence of a large number of pre-existing nodule structures may allow this rapid response.
Based on our results indicating that nodule development was a major target for systemic N signaling a transcriptomics approach has been initiated in Mtr to identify: (i) genes associated with competence to nodulate (“competence program”), i.e. that are turned on or off in nitrogen-starved plants and allow the plant to activate symbiotic signaling with Rhizobia; and (ii) regulatory genes that control early stages of nodule development. The competence program is being analyzed with split root systems, using both wild type and sunn mutant, in which competence to nodulate is not affected by the plant N nutritional status. To identify genes controlling nodule development, we have exploited results of microarray analyses that allowed us to identify 185 regulators of gene expression up-regulated in Mtr nodules. To investigate their involvement in early nodulation steps we have set up a sensitive assay, based on quantitative RT-PCR on 384 plates, to study their expression in a series of wild type and early symbiotic mutants. We were thus able to identify regulatory genes associated with various symbiotic stages, from Nod factor signaling to Rhizobium infection and nodule primordium formation. The next step is to examine the impact of the nitrogen status on the activation of these genes.
As quoted above, N acquisition necessitates a large flow of carbon compounds both at the functional level (for respiration and amino acid synthesis) and for the development of root and nodules structures. As such, below ground structures can compete for assimilates with other plant sinks and carbon supply to nodules could become a limiting factor of N acquisition. In fact, in our experiment we have demonstrated that preferential carbon allocation to the untreated nodulated roots of N-limited plant is a very early event (detected after only 4 days of N deficiency) that may be revealed before any significant change either in N acquisition capacity or in nodule development. This may suggest that such event may be part of the early mechanisms leading to the developmental response.
4 Models for understanding the relationships between plant nitrogen nutrition and plant growth traits
The acquired knowledge has been aggregated in an ecophysiological model that allows simulating the relations between leaf area and N retrieval by roots. This model was developed to explain how nitrate availability in the nutritive solution and the quantity of incident photosynthetic active radiation, two main environmental factors affecting C and N acquisition, affect N retrieval by the plants. Because of the identification by the model of either variables or physiological parameters that could be associated to genetic variability, the model would furnish an analysis grid to characterize and analyze the genotypic diversity of plant N nutrition.
The model has been established using A17 line (cv. Jemalong), for the vegetative period, using data arising from two greenhouse experiments in controlled conditions concerning two environmental factors. The model takes into account symbiotic nitrogen fixation throughout the vegetative period and its modulation by nitrate availability. The proposed model [50] is structured around four integrative variables that characterize plant growth: plant biomass, below ground biomass, amount of plant N and projected leaf area. These integrative variables were decomposed in simpler intermediates variables allowing to hierarchy and quantify the impact of the environmental factors studied on plant growth. These intermediates variables are predicted by the model using response curves to the environmental factors.
The analytical approach described herein has allowed characterizing the adaptative strategy of the plants in response to both nitrogen limited nutrition and carbon limited nutrition. Under nitrogen limited conditions plants reduced their rate of structure set up (leaf area) while plant functioning was little affected. Interestingly a major impact of C nutrition was observed on model parameters driving both structure set up (leaf area and below ground biomass) and functioning (conversion of incident photosynthetic active radiation in biomass, specific N retrieval and efficiency of N conversion to projected surface), suggesting a strong adaptative response to carbon limited nutrition. This very significant result demonstrates clearly that N nutrition cannot be analyzed independently from the interaction between nitrogen and carbon nutrition [51]. At the plant level, when plants are N deficient, they still maintain their specific photosynthetic activity (i.e. carbon dioxide assimilation per unit of foliar surface) but they reduce their leaf expansion. This hence constitutes a relevant phenotypic target for detecting any contrasted N nutrition among various genotypes.
The model provides valuable support for assisting the detection of genetic variants affected for nitrogen uptake, including soil mineral nitrogen retrieval by the roots and symbiotic N2 fixation by root nodules. The key variables (in particular leaf area) identified in the preceding steps were used as criteria for ranking genotypes according to their diversity in their capacity to retrieve N from different sources in a recombinant inbred lines population (LR4 Jemalong X DZA315.16 consisting of 175 inbred lines of Mtr), grown with contrasted nitrate availabilities in the nutritive solution. Plant leaf area was measured at a given date to discriminate genotypes according to their capacity to take up N while the estimation of their nitrogen nutrition index [52] allowed checking if the genotypes were effectively differentially taking up nitrogen. Leaf area at a given date was not a robust trait to highlight differences in N uptake among the genotypes. When measured dynamically, leaf area allowed calculating relative expansion rate which was found to be a good indicator for ranking genotypes according to their ability to uptake nitrogen in given environmental conditions affecting carbon and nitrogen acquisition [51] (Fig. 4).
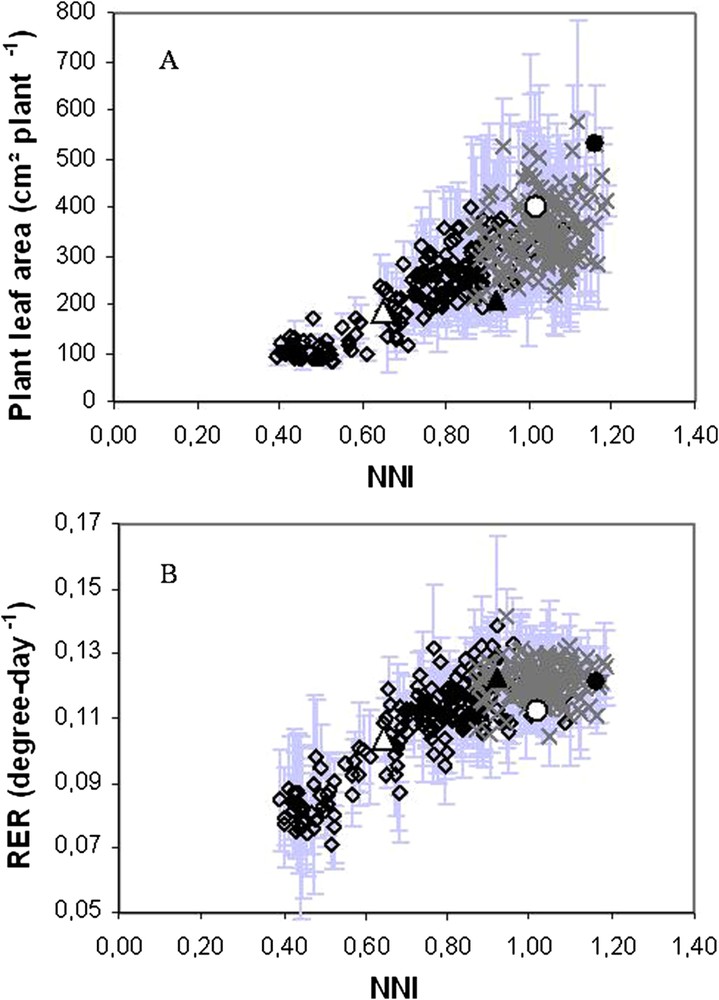
Expressions of plant leaf area as a function of N nutrition index (NNI). In both graphics, each value indicates mean ± SD of three replicates for RILs at 0.625 mM (diamonds) and 10.5 mM of nitrates (crosses). Parental lines are also represented: Jemalong (triangles) and DZA315-16 (circles) for 0.625 mM (opened symbols) and 10.5 mM of nitrate (closed symbols). In (A), plant leaf area is expressed as absolute values at a given date. Leaf area reflects the nitrogen nutrition level of a population of recombinant inbred lines (RILs) issued from the cross between Jemalong and DZA315-16. In (B), plant leaf area was dynamically measured and expressed as relative expansion rate (RER) and so integrates plant growth. At a given time RER was determined as the local slope of the relationship between the logarithm of leaf area (A) and thermal-time. RER was correlated to the nitrogen nutrition level when RILs were affected for N uptake while it was stable under supra optimal N nutrition. From Moreau et al. [52].
This model was also a valuable tool for comparing the functional response to variations in N availability of a root architecture mutant (TR185) with its wild type Jemalong (WT). The mutant TR185, displaying highly branched roots and scarce small nodules phenotype, was selected after γ-ray mutagenesis on Jemalong [32]. In our study, whatever the regimes, the mutant TR185 displayed a higher number of lateral roots than WT but its leaf area was depressed (Fig. 5, A–B). Using the previous model, it was demonstrated that (i) although the mutant has a reduced growth (leaf area) as compared with WT, both genotypes had similar C assimilation and partitioning of biomass to roots; (ii) the mutant and WT had differential N acquisition per amount of root biomass (data not shown) but similar efficiency of N conversion to leaf area (Fig. 5C). As such, the model helped to conclude that the two genotypes contrasted for their root structure only differed functionally in their N acquisition capacity per amount of roots. In parallel with what was described for Sunn, a comparative transcriptomics study between TR185 and WT is underway to better target the genes to be used in reverse genetic strategies and create a new valuable genetic variability.
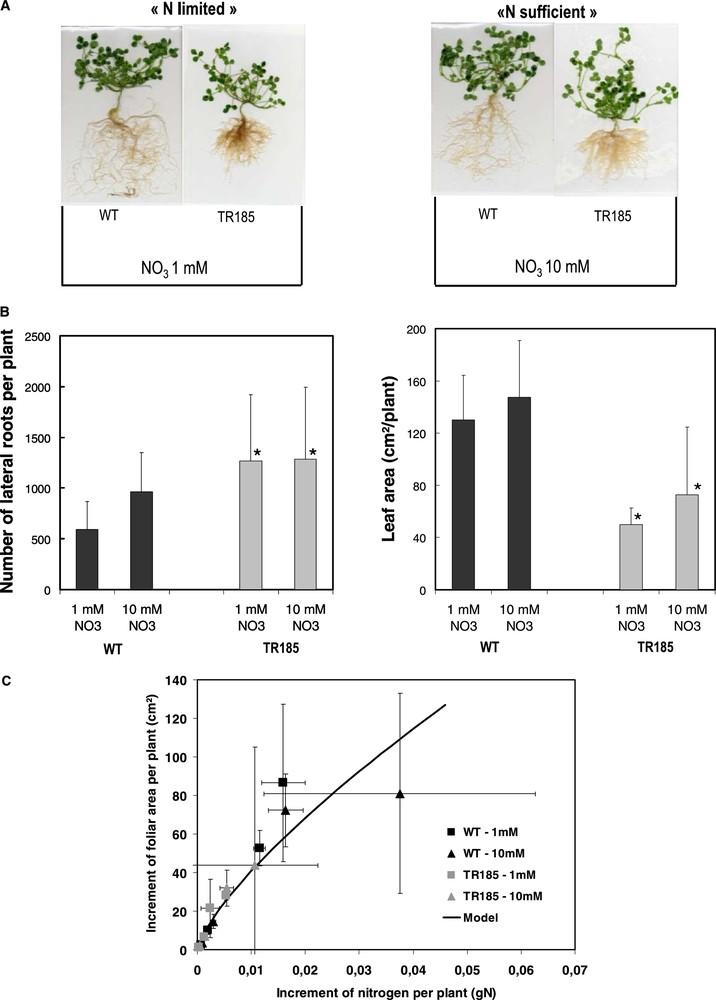
The branched root TR185 mutant was obtained from wild type Jemalong (WT), after γ-ray mutagenesis following the procedure previously described [32]. After germination, plants were grown in a climatic chamber on aerated hydroponic system using a Rhizobium-free nutrient solution containing either 1 or 10 mM of NO3. Plants were sampled and measured for root and shoot traits after 7, 10, 14, 21 and 28 days of growth. (A) Pictures of whole plants 28 days after sowing are presented. (B) At 28 days, number of lateral roots (total of 1st and 2nd order laterals) and leaf area per plant are given for each genotype and treatment. The values are the means of three replicates of one biological repeat and vertical bars indicate SD. *Significant difference according t-test (P < 0.05). (C) The increment of foliar area between two sampling dates was plotted against the increment of N accumulation between two sampling dates and compared to the model curve previously developed [52].
Therefore, the model developed furnishes a grid to analyse a spontaneous or induced genetic variability for plant N nutrition and to detect valuable candidate genes which may improve N acquisition and conversion efficiency, by dissecting very integrative phenotypic variables into more simple variables. In parallel with what was described for Sunn, a comparative transcriptomics study between TR185 and wild type Jemalong is underway to better target the genes to be used in reverse genetic strategies and create a new valuable genetic variability. The model proposed in this work could so constitute an innovative tool for diagnosing the differential origin between lines in their capacity to retrieve N [52].
5 Conclusion
This study was conducted using various expertise at different levels: nodule development, regulation of and absorption, genetic of nodulation, acquisition of N and C and its modeling. Mechanistic molecular approaches were combined with genetic and ecophysiological approaches to characterize the physiological process involved at the level of the plant. A special attention was given to integrative mechanisms which allow adjusting nodulated root functions with the nitrogen demand of the plants.
In the literature, many studies analyzed the legume N2 fixation reaction to elevated mineral nitrogen, whereas the plant adaptation to N source deprivation, a stress with probable occurrence in agronomic situations, has received much less attention. This knowledge is particularly needed for agronomic legumes because of its potential impact on their yield and stability.
Our work help to demonstrate that nitrogen acquisition capacity is adjusted by the N nutritional status of the plant through systemic signaling. We unraveled the components of the adaptative strategy of legume plants faced to modulation of N availability: it occurs on the short term in the case of nitrate fed plants (via both functional and developmental responses) and on the longer term (with a crucial role of nodule organogenesis as target of the plant N status) with N2 fixing plants. It was shown that C metabolites allocated to “compensating” roots (i.e. those still exposed to the N resource upon N starvation on localized part) was very early and instrumental of the plant adaptative response, far before any functional or developmental regulation could be detected.
The use of ecophysiological and genetic tools allows characterizing the physiological processes involved and the underlying molecular determinants. Such tools allow to formalize through modeling the strategy of the plant response to its environment and to identify the molecular basis of genes involved in the functions described by the model. It can also lead to the formulation of modeling tools for high throughput phenotyping. Transcriptomics analysis helped identifying genes whose expression depend upon plant N status. Such genes could so constitute markers indicators of the plant N status and help in phenotyping on large plant population early adaptative responses to situations under which plant encounters N shortage.
The work here presented demonstrates that ecophysiological tools help to characterize a genetic diversity related to plant N nutrition. The first version of the model is based on trophic parameters related to the C and N acquisition and allocation. Root and shoot architecture variables should now be implemented in this model. Ultimately, the model may also integrate the effect of some major genes involved in N nutrition incorporating the contribution of particular alleles.
On the basis of germplasm collection genotyping, or RNAi or TILLING strategies, modelling work offers an innovative strategy to guide the enlargement of the genetic variability for particular candidate genes. It may finally help to propose target ideotypes and valuable alleles to breeders.


