1 Introduction
Ancient species of olive trees (Olea europaea L.), growing near archaeological sites and heritage buildings contribute to the fascination of the Orcia Valley. Centuries ago, these trees were selected and cultivated for their oil. Interesting studies have been conducted to investigate the nutritional properties of oil produced in the valley [1]. The survival of the ancient olive trees was due more to their symbolic than their nutritional value. Recognition of olive cultivars is difficult due to the wide variability of germplasm selected over the centuries by adaptation to microclimate, parasites, cultivation techniques and local production methods.
Studies of the Orcia Valley, combined with historical sources can provide insights into the relationships between archaeological sites and ancient cultivation of olive trees. In particular, the historical presence of cultivated olive trees in the area is said to go back to the seventh century AD, when Demaratus, father of Luchmon, fled from Corinth with riches, men and products that he sold in Etruria. Luchmon later became the fifth legendary king of Rome (616–578 BC) under the name of Lucius Tarquinius Priscus [2].
Remote sensing data can be useful for reconstructing archaeological landscapes. Archaeological surveys in the Orcia Valley did not discover amphoras but dolia of Etruscan and Imperial Roman age, indicating production of oil for local consumption [3,4].
Olive trees are usually characterized pomologically, which involves comparing the trees, leaves and fruits (especially the endocarp) with reference collections and obtaining information about the range of morphologically similar plants that could have originated from similar varieties [5]. Morphometrics (non-invasive analysis of endocarp size and shape) provide a model for investigating differences between archaeological and modern olive endocarps [6]. Quantitative analysis based on study of endocarp outline by elliptic Fourier shape analysis discriminates cultivar groups [7].
The morphology and ultrastructure of carpological specimens have recently been studied by light and electron microscopy, and DNA can also be detected in palaeobotanical material by cytochemical staining with 4’, 6-diamidin-2-phenylindole [8,9,10]. Carpological samples can be used for molecular studies, and ancient olive pits can be used to find ancient DNA [11].
The olive is a prevalently allogamous species with high levels of heterozygosis and polymorphism in apparently similar individuals [12]. The genetic heritage and the numerous synonyms and anonyms have been studied in ancient varieties of olive by collecting material (fruits and leaves) and applying nuclear microsatellite markers [13]. Olive trees have also been the subjects of phylogeographic and phylogenetic studies [14], to develop and evaluate consensus chloroplast primer pairs with highly variable sequence regions in a diverse array of plant taxa [15]. Plastid DNA is generally transmitted via the maternal line and is found in tegument of pits, the only structure that resists aerobic deterioration in archaeological sites [10]. The geographical distribution attained by organelle molecular markers provides a better scenario of past migration history than nuclear markers due to their uniparental mode of inheritance [16].
The aim of the present study was to investigate ancient olive trees growing near archaeological sites and heritage buildings in the Orcia valley, Tuscany, using traditional pomology studies of the trees, fruits and leaves. Morphometric study of size and shape and quantitative analysis of endocarp outline by elliptic Fourier analysis provided a model for discriminating cultivars on the basis of endocarp features. Ultrastructural characterization of endocarp tegument morphology was useful to detect plastid organization in the endocarp and locate DNA fluorescence in plastids of tegument tissues. Molecular studies with cytoplasmic markers were conducted to test similarity and if possible detect variations between individuals. By multidisciplinary comparison of pomological descriptions, morphometric analysis, ultrastructural observations and molecular data, we endeavored to characterize a restricted group of trees.
2 Materials and methods
2.1 Sampling
The plants chosen for sampling (Fig. 1) were the oldest in the area (Mediterranean bioclimatic stage at 43° 10′ N, 11° 30′ E: mean annual maximum temperature 17.4 °C, mean annual minimum temperature 9 °C and mean annual rainfall 750 mm). Records of the landowners of Sesta and Palazzo showed that new olive trees were planted in 1850 and 1870, but the cultivars are not mentioned. The approximate age of the other specimens sampled was evaluated by considering trunk and stump prominences and bark consistency (Fig. 2). Trees were labeled with non-toxic gold paint for subsequent recognition. The sampling areas were identified topographically and spatially by GPS recording (Table 1). For analysis, mature fruits and adult leaves were sampled in November seasons 2009 and 2010. Samples were obtained using tweezers, gloves and sterile containers, which were numbered and catalogued.
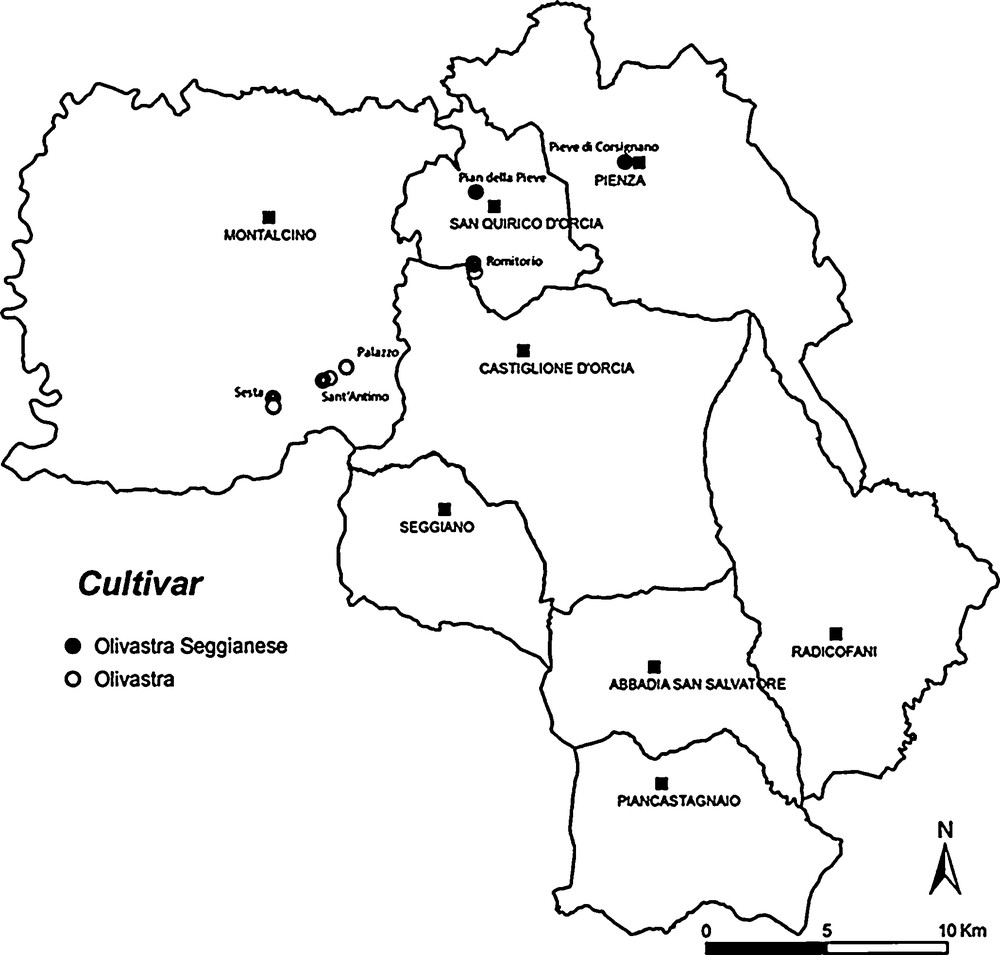
The sampling sites of ancient trees in the Orcia Valley.

St. Antimo monastery and olive trees growing close to the facade.
Description of sampling sites, GPS coordinates, and characteristics of trees, fruits, endocarps and leaves.
| Place | GPS | Trunk circumference (m) | Cultivation method estimated age | Fruit polar and transverse diameter (mm) | Fruit average weight (g) | Endocarp perimeter (mm) and surface area (mm2) | Endocarp polar and transverse diameter (mm) | Major axis sterile and fertile valva (mm) | Distance base-centroid (mm) | Endocarp average weight (g) | Leaf width and length (mm) |
| 1. Romitorio 1 | 43°2’16” N 11°35’34” E |
2.9 | Wild 1900 | 17.5 15 |
2.7 | 32.9 75.83 |
11.56 7.888 |
3.33 4.554 |
5.641 | 0.43 | 13.5 64 |
| 2. Romitorio 2 | 43°2’17” N 11°35’33” E |
2.6 | Wild 1900 | 16 15 |
2.5 | 32 72 |
10.87 8.020 |
3.51 4.536 |
5.193 | 0.45 | 13.3 64 |
| 3. Sesta 1 | 42°59’30” N 11°29’16” E |
3.6 | Cultivated 1850 | 17.7 15.7 |
2.82 | 32.46 71.24 |
12.26 7.383 |
3.484 3.9 |
9.934 | 0.48 | 14.5 73 |
| 4. Sesta 2 | 42°59’31” N 11°29’17” E | 3.8 | Cultivated 1850 | 17.5 15.5 |
2.80 | 31.87 78.55 |
11.42 7.912 |
3.272 4.64 |
5.65 | 0.47 | 15.5 74 |
| 5. Palazzo 1 | 43°0’14” N 11°31’37” E |
3.55 | Cultivated 1870 | 17.5 15.4 |
2.55 | 31.78 66.22 |
11.98 7.378 |
3.034 4.344 |
5.88 | 0.48 | 12 54 |
| 6. Palazzo 2 | 43°0’13” N 11°31’36” E |
3.8 | Cultivated 1870 | 17.5 15.5 |
2.57 | 33.7 75.92 |
12.39 7.5 |
3.5 4 |
5.925 | 0.49 | 12 54 |
| 7. S. Antimo 1 | 42°59’59” N 11°30’55” E |
5.1 | Cultivated 1800 | 17.5 15 |
2.72 | 32.22 73.85 |
11.56 7.917 |
3.367 4.55 |
5.868 | 0.57 | 13.4 63 |
| 8. S. Antimo 2 | 42°0’0” N 11°30’59” E |
7.35 | Cultivated 1750 | 17.4 15.3 |
2.50 | 31.78 69.42 |
12.13 7.546 |
3.728 3.818 |
5.837 | 0.49 | 12.5 54 |
| 9. Pian della Pieve | 43°4’5” N 11°35’41” E |
2.43 | Cultivated 1910 | 17.8 15.7 |
2.83 | 31.92 71.03 |
11.55 7.975 |
3.583 4.394 |
5.946 | 0.50 | 13.1 62 |
| 10. Pieve di Corsignano | 43°4’38” N 11°40’16” E |
2.8 | Wild 1890 | 17.5 14.5 |
2.4 | 30.75 66.1 |
11.40 7.915 |
3.42 4.490 |
5.675 | 0.4 | 14 66 |
2.2 Traditional pomology
The traditional pomology table was drawn up according to the International Olive Council (IOC) on the basis of the Word Catalogue of Olive Varieties [17] and autochthonous reference collections in the herbarium of Siena University Botanic Gardens. To have enough detail, we determined the traditional characters of trees, leaves and fruits (listed in Table 1). Observations by stereomicroscopy were conducted on adult leaves obtained from the median part of 1-year-old branches. The mean length and width about of 100 leaves was recorded for each sample. The fruit observations were conducted on a mean about of 220 fresh mature samples. We determined the transverse and polar diameters and weight. The epicarp and mesocarp were removed and morphometric studies (size and shape analysis) were conducted on the mature endocarp.
2.3 Morphometric size and shape analysis
To analyze endocarp structure (Fig. 3.1) and for specific non-invasive morphometric study [6], mature endocarps were gently but thoroughly cleaned with a plastic brush. The ideal average sizes of about 220-270 olive endocarps per tree are illustrated in Tables 1 and 2. The morphometric study of size by stereomicroscope considered endocarp perimeter and area, polar and transverse diameters, total major axis of sterile and fertile valves and base-centroid distance (Table 1). Measurements were made by digital micrography on endocarp images imported from files and processed with the public domain ImageJ programme (CyberMetrics) [18]. Artifacts were reduced by subtracting background and graticule image calibration using plug-ins of the ImageJ program. Errors of measurement due to samples of uneven size were adjusted by adding more 40–50 mature specimens. The average size of the scalar profile of sterile and fertile valves of Table 2 were obtained from the many digital images processed by ImageJ as described by Terral et al. [6], modified: each valve of each endocarp was identified from the suture line, which was then taken as base. Then single valves were rotated and placed with the base on the x-axis and 19 pseudohomologous reference points (x, y) were recorded between the two homologous points at (0, 0) and (0, 1) defining the base and apex of the valve. The homologous and pseudohomologous data obtained was processed using Microsoft Excel for Windows to reproduce the harmonic profile for each valve.
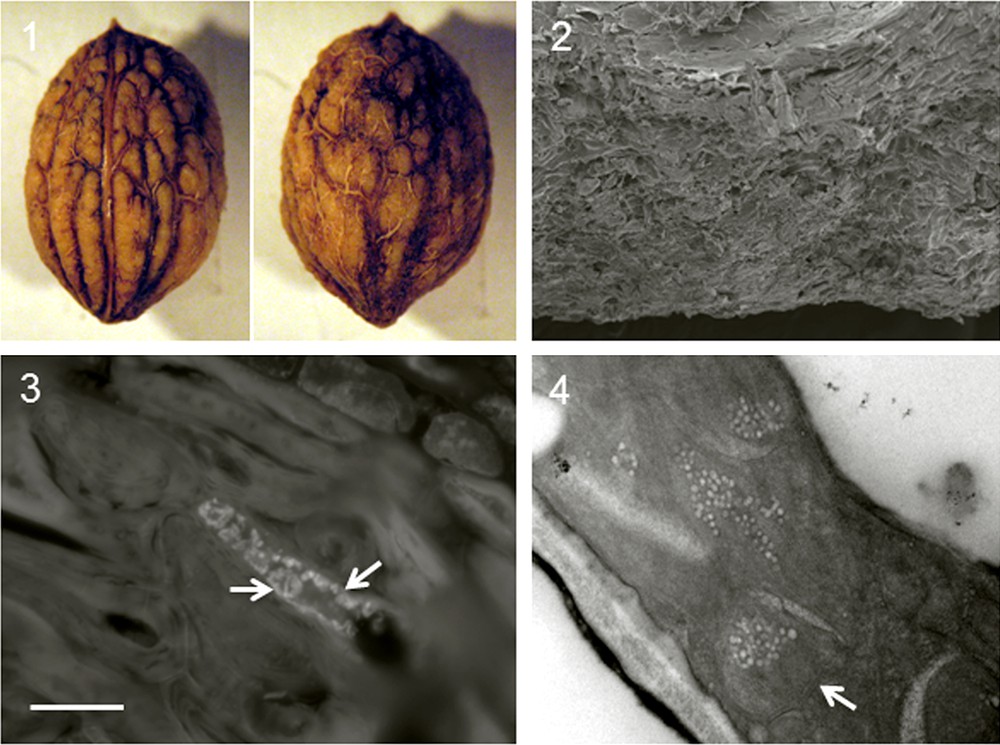
Micrographs – bar at lower left of 2.3 is valid for all micrographs of Fig. 2. 3.1 Light microscope images of olive endocarps showing morphological structure: lateral (with suture separating flatter sterile valve on left from fertile valve on right) and dorsal. Endocarp carpellar fascicles, base and apex are clearly visible. Bar = 3.77 mm. 3.2 Scanning electron micrograph of tegument cells showing disorderly, irregular distribution. Bar = 250 μm. 3.3 Fluorescence microscope image with DAPI showing well-conserved plastids containing high-contrast DNA (arrows) in central part of tegument cell; bar = 1250 μm. 3.4 Transmission electron micrograph showing tegument ultrastructure with well preserved plastids (arrows). Bar = 2500 μm.
Homologous and pseudohomologous values for reconstruction of scalar profiles of sterile and fertile valves. The sterile (S V) and fertile (F V) valves of each endocarp were placed on the x-axis and 19 pseudohomologous reference points (Y), intermediate between the two homologous points (0, 0) and (0, 1) constituting the valve base and apex, were recorded. The major axis of sterile and fertile valves is indicated in bold.
| 1. Romitorio 1 | 1. Romitorio 1 | 2. Romitorio 2 | 2. Romitorio 2 | 3. Sesta 1 | 3. Sesta 1 | 4. Sesta 2 | 4. Sesta 2 | 5. Palazzo 1 | 5. Palazzo 1 | 6. Palazzo 2 | 6. Palazzo 2 | 7. S. Antimo 1 | 7. S. Antimo 1 | 8. S. Antimo 2 | 8. S. Antimo 2 | 9. Pian della Pieve | 9. Pian della Pieve | 10. Pieve di Corsignano | 10. Pieve di Corsignano |
| V S | V F | V S | V F | V S | V F | V S | V F | V S | V F | V S | V F | V S | V F | V S | V F | V S | V F | V S | V F |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1,15 | 0,9 | 1,7 | 2 | 0,9 | 0,7 | 1 | 1 | 1,1 | 0,7 | 1,5 | 0,9 | 1,7 | 1,7 | 1,2 | 1 | 1,6 | 1,1 | 1,4 | 1 |
| 2 | 2,1 | 2,5 | 2,6 | 1,7 | 2,2 | 2,1 | 2,2 | 1,5 | 1,8 | 2 | 2,2 | 2,5 | 2,3 | 2 | 2,3 | 2,5 | 2,2 | 2 | 2,2 |
| 2,6 | 2,7 | 3,1 | 3,2 | 2,5 | 2,9 | 2,6 | 3 | 2,2 | 2,4 | 2,6 | 2,7 | 2,8 | 3,2 | 2,6 | 2,7 | 2,9 | 2,8 | 2,6 | 3 |
| 3 | 3 | 3,3 | 3,6 | 3,1 | 3,3 | 2,9 | 3,4 | 2,6 | 2,7 | 2,9 | 3,2 | 2,9 | 3,5 | 3,1 | 3,2 | 3,1 | 3,5 | 3 | 3,5 |
| 3,2 | 3,3 | 3,4 | 3,7 | 3,2 | 3,5 | 3,1 | 3,7 | 2,85 | 3 | 3,2 | 3,6 | 3 | 3,8 | 3,4 | 3,3 | 3,3 | 3,7 | 3,2 | 3,9 |
| 3,25 | 3,5 | 3,5 | 4 | 3,3 | 3,7 | 3,2 | 4 | 2,9 | 3,3 | 3,3 | 3,8 | 3 | 4,1 | 3,55 | 3,5 | 3,35 | 4 | 3,3 | 4,2 |
| 3,33 | 3,7 | 3,51 | 4,1 | 3,4 | 3,7 | 3,25 | 4,2 | 3 | 3,8 | 3,45 | 4 | 3,1 | 4,2 | 3,6 | 3,6 | 3,4 | 4,1 | 3,4 | 4,3 |
| 3,25 | 3,8 | 3,51 | 4,2 | 3,37 | 3,75 | 3,27 | 4,4 | 3 | 4 | 3,45 | 3,95 | 3,2 | 4,3 | 3,7 | 3,7 | 3,5 | 4,15 | 3,4 | 4,3 |
| 3,2 | 4,1 | 3,5 | 4,4 | 3,35 | 3,8 | 3,25 | 4,5 | 2,9 | 4,34 | 3,45 | 3,95 | 3,25 | 4,3 | 3,72 | 3,7 | 3,4 | 4,2 | 3,42 | 4,3 |
| 3,1 | 4,3 | 3,4 | 4,5 | 3,35 | 3,85 | 3,25 | 4,55 | 3 | 4,3 | 3,5 | 3,95 | 3,3 | 4,4 | 3,7 | 3,75 | 3,4 | 4,25 | 3,42 | 4,4 |
| 3 | 4,4 | 3,3 | 4,53 | 3,35 | 3,9 | 3,24 | 4,6 | 3,03 | 4,25 | 3,5 | 3,9 | 3,36 | 4,45 | 3,65 | 3,81 | 3,4 | 4,35 | 3,4 | 4,49 |
| 2,9 | 4,55 | 3,3 | 4,5 | 3,3 | 3,9 | 3,22 | 4,64 | 3 | 4,2 | 3,45 | 3,8 | 3,3 | 4,5 | 3,6 | 3,8 | 3,4 | 4,35 | 3,35 | 4,4 |
| 2,75 | 4,5 | 3,2 | 4,3 | 3,2 | 3,9 | 3,1 | 4,6 | 2,9 | 4,1 | 3,4 | 3,7 | 3,2 | 4,55 | 3,5 | 3,75 | 3,2 | 4,39 | 3,3 | 4,4 |
| 2,65 | 4,2 | 3 | 4,1 | 3 | 3,8 | 3 | 4,5 | 2,8 | 4 | 3,3 | 3,5 | 3,15 | 4,4 | 3,4 | 3,7 | 3 | 4,3 | 3,2 | 4,2 |
| 2,4 | 4 | 2,9 | 3,9 | 2,8 | 3,6 | 2,9 | 4,3 | 2,6 | 3,8 | 3,1 | 3,3 | 3,05 | 4,2 | 3,1 | 3,5 | 2,8 | 4,2 | 3 | 4 |
| 2,1 | 3,85 | 2,7 | 3,6 | 2,4 | 3,2 | 2,7 | 4,1 | 2,5 | 3,5 | 2,9 | 3 | 2,5 | 4 | 2,8 | 3,3 | 2,6 | 3,9 | 2,7 | 3,6 |
| 1,6 | 3,45 | 2,4 | 3,3 | 2 | 2,7 | 2,2 | 3,5 | 2,2 | 3 | 2,2 | 2,7 | 2,1 | 3,8 | 2,5 | 2,9 | 2,2 | 3,6 | 2,2 | 3,2 |
| 1 | 2,5 | 1,9 | 2,7 | 1,4 | 1,8 | 1,8 | 3 | 1,7 | 2,5 | 1,7 | 2,3 | 1,3 | 3,3 | 1,8 | 2,3 | 1,7 | 3,2 | 1,6 | 2,2 |
| 0,7 | 1,9 | 1,1 | 2 | 0,4 | 1 | 0,8 | 1,7 | 1 | 1,3 | 0,7 | 1,8 | 0,8 | 2,6 | 1 | 1,6 | 1,1 | 2,5 | 1 | 1,5 |
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
For shape analysis, about 20 average harmonic olive stones were selected from 220–270 endocarps for each cultivar. Each endocarp was disposed on an illuminated dishboard to acquire the perfect profile. The samples were placed accurately on their lateral side with the apex pointing up, the suture line well visible along the vertical axis and the fertile valve on the right. The images (JPEG format, 24 bit) were acquired with a high-resolution digital camera, Nikon Coolpix P6000 (13.5 real MP) with good macro features and optical × 4 Nikkor lens. A metric mark was placed near each group of endocarps and used as reference to record size. An automated thresholding procedure was performed by the Otsu method [19], which detects proper gray-level edges. Each image was primarily binarized and a total of 180° points (x, y), at equal angular intervals from the centroid (one point every 2°), were digitized along the outlines using a Matlab procedure (rel. 7.1; PLSToolbox Eigenvectorb 4.0). The overall shape of samples was obtained by Elliptic Fourier Analysis (EFA) on the outline coordinates [20]. The outline can be approximated by a polygon of x–y coordinates. EFA is based on the separate Fourier decompositions of incremental changes in x and y coordinates as functions of the cumulative chord length of the outline polygon. It yields the spectrum of sample shape closed contours in terms of harmonically related trigonometric curves. For each harmonic equation, two Fourier coefficients are computed for the x and y projections, so that the total number of coefficients is 4n, where n is the number of harmonics fitted to the outline [21]. The total number of harmonics that can be computed for any outline is equal to half the total number of outline coordinates (the “Nyquist frequency”). The Fourier series was stopped at value k where the average cumulative power was 99.999% of the average total power [7]. For any outline, the total power was calculated as the sum of individual harmonic powers from 1 to k, where k is the Nyquist frequency [21]. The harmonic coefficients describe the size, shape and orientation of each harmonic ellipse and are analyzed by multivariate statistics. The elliptic Fourier coefficients were normalized to be invariant with respect to size, location, rotation and starting position according to Rohlf [20]. Cartesian coordinates were used. EFA and all further analyses [22] were performed using Matlab 7.1 (The MathWorks, Natick, USA). Mean outline and standard deviation ranges were reported graphically for each cultivar. Euclidean distances between mean shape configurations of each cultivar were calculated with the complete linkage algorithm [23], using the harmonic EFA coefficients. Results were plotted as a cladogram.
2.4 Morphological and ultrastructural study of endocarp tegument
Endocarps were selected for morphological and ultrastructural study on the basis of similar pomological descriptions (Table 1). The method described in Milanesi et al. [10] was used to determine tegument ultrastructure of the fresh endocarp and to locate DNA in tegument tissue by cytochemical stains.
2.5 Molecular analysis of endocarp tegument
To examine intra-population variability, we monitored haplotypes of O. europaea L. by amplification of plastid microsatellites identified in the literature and GenBank (http://www.ncbi.nlm.nih.gov) on the basis of high mononucleotide variability (n ≥ 7), preferring those with high polymorphism. A crucial point for molecular characterization was amplification of DNA fragments of less than 120 bp by PCR [24,25]. Molecular study was conducted in a first phase to define the total DNA extraction protocol and verify similarity between endocarp tegument and leaves of the same individual using two SSR loci (NTCP9 and ccSSR8). The study was extended to endocarp tegument of the other samples using loci such as ccSSR5, ccSSR7, ccSSR18, ccSSR19 and ccSSR20 [26], which meet the above criteria. Allelic sizing was obtained with MegaBACE™ 1000 DNA analysis system (GE Healthcare Life Science) and evaluated using Fragment Profiler version 1.2 (GE Healthcare Life Science), following known lab protocols [27].
3 Results and discussion
3.1 Sampling
The sampling areas in the Orcia Valley were identified from historical and remote sensing data (Fig. 1), and ancient trees growing near heritage buildings were preferred (Fig. 2), because olives are a symbol of peace for the Christian community and those growing near monasteries were cultivated more for their symbolic value than for production. These plants are also interesting because of their longevity, having survived climatic and parasitic adversities and having adapted to changes in cultivation techniques. They were therefore judged the most suitable plants on which to attempt identification of the original cultivars of the Orcia Valley, selected by the local communities on an empirical basis. They were probably propagated by agamic multiplication from a progenitor and would therefore have distributions limited to specific areas. The trees sampled in the present study had an age between 100–110 and 250 years (Fig. 1). Apart from those at Sesta and Palazzo, for which we have documentation on the date of planting, the stumps of others could be older, because they can regenerate if the tree is damaged. Olive trees do not form true annual growth rings and their dimensions can be influenced by environmental factors. It is therefore difficult to date them accurately except by radiocarbon wiggle matching [28]. Since this method requires fragments of trunk heartwood, it is unsuitable for monumental trees.
3.2 Traditional pomology description
For the preliminary pomology table we recorded characters of trees, fruit (epicarp, mesocarp and endocarp) and leaves (Table 1). Analysis of the fruit was considered useful for the subsequent aims of identifying and describing olive germplasm of the Orcia Valley from endocarps found in archaeological sites [6]. Regarding pomology graphic values, we observed the range of cultivars with suitable morphological characteristics. They were ancient trees, tall and erect, with thick spreading crowns, high vigor and limited distribution, resistant to freezing in the hill-belt climate (e.g. winter 1956 and 1985) and to parasites. Traditional biometric parameters of fruit mainly concerned qualitative and quantitative variations in the surface of the epicarp and endocarp, and showed few morphological differences. The color of the epicarp was black; its form was small, roundish and symmetrical. A single epicarp weighed 2.15–2.85 g. Structure was small, ovoid, symmetrical, with a corrugated surface and many deep, irregular vascular furrows (Fig. 3.1). A single endocarp weighed 0.37–0.57 g. The weight of ripe fruit presumably varied in relation to tree vigor due to pruning and other care. We found that leaf morphology varied in relation to age, position and vigor of the branches, and therefore depended on tree care. To limit these variations, leaves were sampled from the medial part of 1-year-old branches. Adult leaves were elliptical lanceolate and of intermediate size, with a length of 54–74 mm and a width of 12–15.5 mm.
When compared with reference collections [17,29,30], ancient tree morphology, traditional phomology of endocarps and leaves were compatible with olive varieties such as “olivastra Seggianese” cultivated in the Orcia Valley.
3.3 Endocarp morphometric study size and shape analysis
Tables 1 and 2 show endocarp size data analysis. The approach made it possible to characterize the size of a variable morphological structure by means of well-defined parameters. Morphological variability in contemporary cultivars reveals phenotypic divergence in groups of individuals diversified by climatic, ecological, historical and sociocultural factors [6]. Morphometric analysis of endocarp size showed an irregular surface with carpellar fascicles separated by a suture line having a mean width of about 0.2–0.3 mm. The sterile valve (SV) is normally flatter than the fertile valve (FV). The scalar profile of the sterile and fertile valves was recorded as 21 reference points (2 homologous and 19 pseudohomologous), as shown in Table 2. Excel software was used to process the columns S V and F V to obtain line drawings of valve profile Mean polar diameter of endocarps was < 12 mm for Sesta 1 and S. Antimo 2, 11.5 mm for Romitorio 1, Sesta 2, S. Antimo 1, Pian della Pieve and Pieve di Corsignano, and ≥ 10.8 mm for Romitorio 2. Mean transverse diameter of endocarps ranged from 7.3 to 8 mm; the group of endocarps from Romitorio 1, Sesta 2, S. Antimo 1, Pian della Pieve and Pieve di Corsignano had a mean transverse diameter of about 7.9 mm. Endocarp size analysis did not provide a discriminant for determining olive cultivars [6], however average size study enabled reconstruction of ideal valve profiles and calculation of ideal average transverse and polar diameters and ideal shape of the endocarps of each ancient tree. This made it possible to identify a distinct group of olive endocarps (Romitorio 1, Sesta 2, S. Antimo 1, Pian della Pieve and Pieve di Corsignano) of homogeneous proportions with values that describe a very curved scalar profile and morphology characterized by a relatively small polar diameter and a relatively large transverse diameter (Table 2).
Shape analysis: the correct number of harmonics to use for computing the lateral profile of endocarps was calculated on all samples (“Nyquist frequency” = 90). The value selected (i.e. the first value exceeding 99.999%) was 20 harmonics. The mean configuration (black line) and standard deviation (gray lines) for each cultivar are shown in Fig. 4. It is possible to observe distinct shapes belonging to the different cultivars. Romitorio 2, Sesta 1, Palazzo 1 and 2 and S. Antimo 2 have a sharp apex, while Romitorio 1, Sesta 2, S. Antimo 1, Pian della Pieve and Pieve di Corsignano have an ovoid shape with a convex apex. The cladogram, built on Euclidean distances between the mean shape configurations of each cultivar, calculated with the complete linkage algorithm using the EFA harmonic coefficients, appears in Fig. 5. At a linkage distance of 0.06, cluster analysis distinguished three groups. The first included Romitorio 1, S. Antimo 1, Sesta 2, Pieve di Corsignano, Pian della Pieve and San Antimo 2, the second Palazzo 1 and Palazzo 2, and the third Romitorio 2 and Sesta 1. The endocarp shapes of the cultivars of each group generally appeared to be similar.
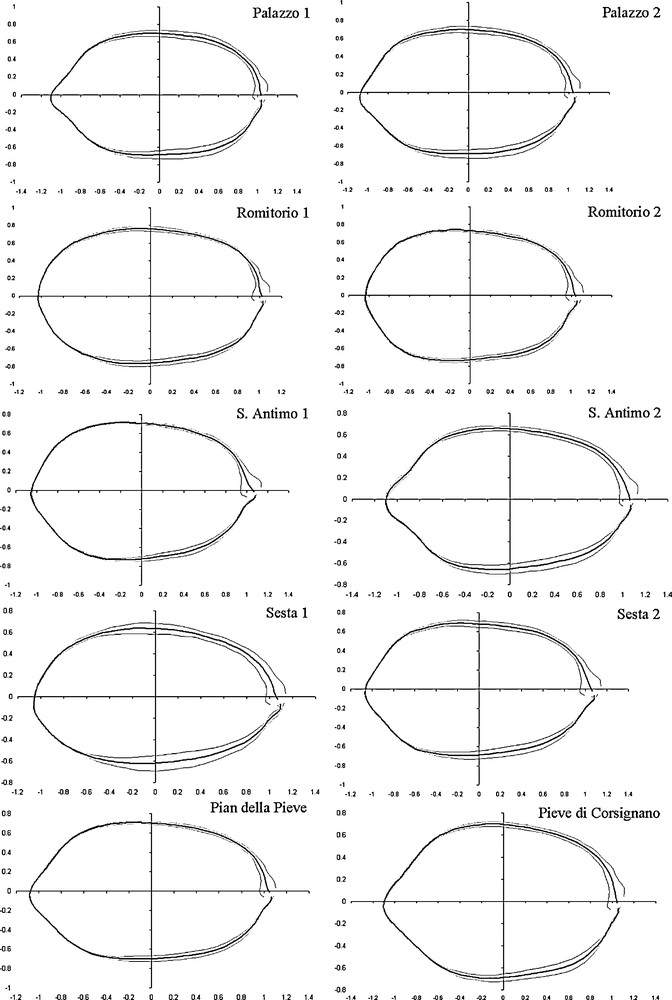
Mean endocarp outline (black line) and standard deviation (gray lines) for each cultivar, extracted with about 20 harmonics.
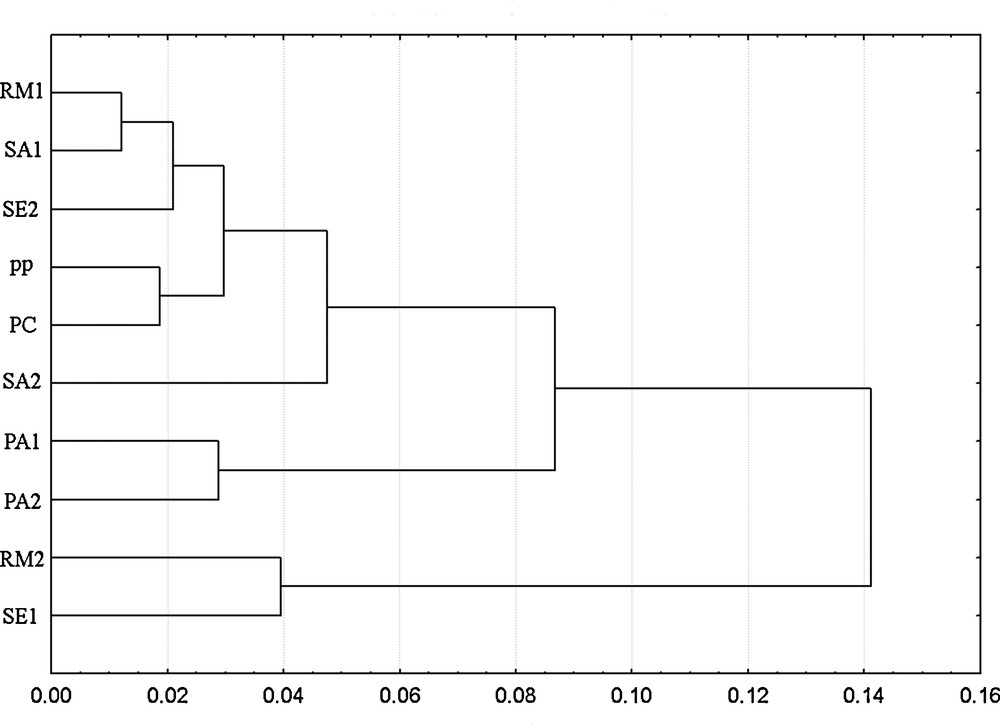
Cladogram based on Euclidean distances calculated with the complete linkage algorithm using the EFA harmonic coefficients extracted in the study. Romitorio (RA), S. Antimo (SA), Sesta (SE), Pian della Pieve (PP), Pieve di Corsignano (PC), Palazzo (PA).
3.4 Morphological and ultrastructural observation
Endocarp tegument structure can conserve useful morphological and genetic information for centuries. To develop effective characterization, in this first phase we processed samples from Romitorio 1, Sesta 2, S. Antimo 1, Pian della Pieve and Pieve di Corsignano for ultrastructural study. The endocarp samples had similar pomological characteristics (shape, size, symmetry, surface features, presence and depth of fibrovascular furrows, base and apex form), a transverse diameter of about 7.9 mm and a polar diameter of about 11.5 mm (Tables 1 and 2). Light and electron microscope observations of endocarp tegument showed morphologically similar samples. Data of the S. Antimo 1 site was therefore taken as representative (Fig. 2); the trees grew within a few meters of the facade of St. Antimo monastery, where Cistercian monks have selected olive germplasm since antiquity, propagating from an ancestor with good resistance to cold and parasites and a high yield of good quality olive oil.
Ultrastructural analysis and cytochemical stains (Fig. 3) enabled assessment of subcellular structure and localization of DNA in endocarp tegument by fluorescent staining. Ultrastructural observation showed that teguments of sterile and fertile valves had the same ultrastructural characteristics. The tegument cells were large and compact with an irregular arrangement imparting mechanical resistance (Fig. 3.2). Plastids were surrounded by a coat containing DNA, presumably prokaryotic. The fluorescent marker 4’, 6-diamindin-2-phenylindole (DAPI) confirmed the presence of DNA in tegument cells (Fig. 3.3): highly fluorescent plastids were observed especially in the central part of tegument cells. The homogeneity of the ultrastructural observations confirmed the results of Besnard and Benvillé [25] who found well-defined groups, little ultrastructural differentiation and low polymorphism of cytoplasmic DNA. The scarcity of tegument plastids was confirmed by transmission electron microscope; this involved the difficult task of sectioning the strong wall of tegument cells, high in lignin, in order to reach the well-conserved plastids in the central part. Plastids were in turn protected by a double membrane (Fig. 3.4 arrow). Thus endocarp organization and tegument ultrastructure composed of irregularly disposed sclereid fibres may favor protection of plastids and the DNA they contain. This is propitious for detecting ancient plastid DNA in carpological remains from archaeological sites.
3.5 Molecular studies
In some cases, environmental conditions that influence plant characteristics and adaptability to microclimates, parasites and local cultivation methods have probably induced changes in pomological, phenological and bioagricultural lines, involving molecular mechanisms and generating polymorphisms in regions of high variability [12]. Nuclear markers are widely used for cultivar identification in olives and other species [31]. They were not considered a priority in the present study because in this preliminary phase we wanted to use cytoplasmic markers to test endocarp characteristics in a limited group of individuals derived from a similar genetic strain, in order to develop a method involving phylogeographical studies [32], to be applied to archaeological endocarps in which it was hoped to detect DNA. We therefore preferred cytoplasmic markers, which the literature indicated to show a high degree of polymorphism; during molecular characterization, this involved amplification of fragments smaller than 120 bp in size [24,25]. Molecular studies were first conducted to extract plastid DNA from endocarps and leaves and to check for similarity using two plastid loci (NTCP9 and ccSSR8) that the literature indicates as functional [26]. We checked genetic similarity and allele identity between endocarps and leaves of the same plant (Table 3), demonstrating a close genetic correlation. We then extended the molecular studies to endocarps of other samples. In this case, comparison with germplasm typical of Tuscan cultivars was not considered a priority because we were concerned with detecting the molecular characteristics of germplasm of the restricted group of ancient olive trees having endocarps with similar morphological characteristics. We checked molecular compatibility with lineages of the Mediterranean geographical area [15,26]. Comparison of the molecular data confirmed that olive trees (Romitorio 2, Sesta 1, Palazzo 1 and 2, St. Antimo 2) showed similar allele profiles, while we found a central group of trees (Romitorio 1, Sesta 2, St. Antimo 1, Pian della Pieve, Pieve di Corsignano) with the same allele (Table 3). The analyses showed low variability among all samples, indicating a close genetic correlation within the restricted group of trees considered.
Comparison of multi-locus profiles of the ten samples of O. europaea L. obtained with seven plastidial microsatellite markers. Comparison of endocarp tegument and leaf samples by two-plastid loci (NTCP9 and ccSSR8) showed within-individual genetic similarity. Romitorio 1, Sesta 2, S. Antimo 1, Pian della Pieve and Pieve di Corsignano (underlined values) showed the same endocarp multi-locus profiles.
| Specimens | 1. Romitorio 1 | 2. Romitorio 2 | 3. Sesta 1 | 4. Sesta 2 | 5. Palazzo 1 | 6. Palazzo 2 | 7. S. Antimo 1 | 8. S. Antimo 2 | 9. Pian della Pieve | 10. Pieve di Corsignano | Leaves 1 Seeds 2 |
| NTCP9 | 183 bp | 184 bp | 183 bp | 184 bp | 184 bp | 184 bp | 183 bp | 183 bp | 183 bp | 183 bp | 1 |
| 183 bp | 183 bp | 183 bp | 184 bp | 184 bp | 184 bp | 183 bp | 183 bp | 183 bp | 183 bp | 2 | |
| ccSSR5 | 1 | ||||||||||
| 84 bp | 84 bp | 84 bp | 84 bp | 84 bp | 84 bp | 84 bp | 84 bp | 84 bp | 84 bp | 2 | |
| ccSSR7 | 1 | ||||||||||
| 120.7 bp | 120.7 bp | 120.7 bp | 120.7 bp | 120.7 bp | 120.7 bp | 120.7 bp | 120.7 bp | 120.7 bp | 120.7 bp | 2 | |
| ccSSR8 | 153 bp | 153 bp | 153 bp | 153 bp | 153 bp | 153 bp | 153 bp | 153 bp | 153 bp | 153 bp | 1 |
| 153 bp | 153 bp | 153 bp | 153 bp | 153 bp | 153 bp | 153 bp | 153 bp | 153 bp | 153 bp | 2 | |
| ccSSR18 | 1 | ||||||||||
| 153 bp | 153 bp | 153 bp | 153 bp | 153 bp | 153 bp | 153 bp | 153 bp | 153 bp | 153 bp | 2 | |
| ccSSR19 | 1 | ||||||||||
| 177.5 bp | 176.5 bp | 176.5 bp | 177.5 bp | 176.5 bp | 176 bp | 177.5 bp | 176.5 bp | 177.5 bp | 177.5 bp | 2 | |
| ccSSR20 | 1 | ||||||||||
| 107 bp | 107 bp | 107 bp | 107 bp | 107 bp | 107 bp | 107 bp | 107 bp | 107 bp | 107 bp | 2 |
4 Conclusions
Remote sensing data and historical research on the Orcia Valley can help understand the relationships between archaeological sites, heritage buildings and ancient olive trees. The trees have natural longevity (100–250 years old) and have survived freezing. They were probably propagated by agamic multiplication from a progenitor and selected by local communities on an empirical basis, as well as for their symbolic value, form and beauty. Generic pomology of endocarps and leaves and specific non-invasive morphometric analysis of size and shape of endocarps of the restricted group of ancient olive trees sampled were closely compatible with the cultivated olive varieties “olivastra Seggianese”. The ultrastructural observations were useful to detect plastid organization in the endocarp and locate DNA fluorescence in plastids of tegument tissues. A close genetic correlation was confirmed on the basis of genetic similarity and identical alleles in endocarps and leaves of the same plant and similar allele profiles in endocarps of the restricted group of ancient olive trees considered. The data acquired by the inverse process we used, starting with the characteristics of archaeological endocarps, could facilitate rapid identification of cultivars such as “olivastra Seggianese”, that originated on Mt. Amiata (Siena). For effective and valid comparison of modern and archaeological endocarps of typical cultivars, difficulty may arise in making the modern material similar to the archaeological material, in the limited number of archaeological samples available, and in the perishability of archaeological genetic material. Rigorous pomological study, development of specific morphometric methods of size and shape analysis for olive cultivars, more efficacious ultrastructural and molecular studies, and the use of correct archaeological methods such as dendrochronology and palinology may lead to better interdisciplinary techniques and new insight useful for more detailed study of archaeological olive pits and for the valorization of typical products, such as olive oil of the Orcia Valley.
Conflict of interest statement
The authors have not declared any conflict of interest.
Acknowledgments
We thank Ariano Buracchi for his able and enthusiastic collaboration, Albero Comini and Andrea Masi (Archaeological Science Department, University of Siena), for help with sampling, Claudia Faleri and Fabrizio Ciampolini for assistance with light and electronic microscopy, Monica Scali for help with molecular analysis and Giampiero Cai for suggestions about available software for size analysis.


