1 Introduction
Wheat is the most widely grown crop in the world, representing a major renewable resource for food, feed, and industrial raw materials. The world demand for food by an ever-growing population is expected to increase, by about 40% in 2030 [1]. Meeting this demand would require that agricultural land stabilizes at ca. 1.5 billion hectares [2] together with an annual yield increase of 2%, while yields have increased by only 0.9% per year over the past decade [1]. The situation is even worse in Europe (the first world producer) and particularly in France, where wheat yields have been stagnating since 1995, presumably because of unfavorable climatic conditions [3]. Domestication and crop improvement events have taken advantage of the genetic diversity of crops and their relatives. Hence, understanding the genetic bases of these events may help us imagine new ways to exploit untapped genetic resources and to accelerate genetic progress through modern breeding methods.
Cereals are amongst the first species to have been domesticated by man, at nearly the same time as dogs, sheep and cattle. Respective domestications occurred independently on all Continents. This paper will focus on the history of wheat, which was one of the first cereals to be domesticated in the Middle East and subsequently spread over the Old World during the Neolithic revolution. For plants, domestication is tte suite of anatomical domestication is the suite of anatomical and morphological changes that follows cultivation being oriented toward an adaptation to the new anthropized environment. In this sense, domestication is different from conscious cultivation, which began with related wild species [4]. Moreover, the discovery of fossilized remains on archeological sites does not allow ascertaining whether they were used for cultivation or for gathering. Molecular genetics tools may reconstruct the evolutionary scenarios of wheat domestication. This review addresses five questions about wheat domestication: why, when, where, how, and what the consequences are for today's agriculture.
2 Wheat is a polyploid series
As early as the late XIXth century, the first studies on biogeography of cultivated crops allowed identifying specific regions called the centers of origin [5,6]. For wheat, barley, pea and lens this center of origin is located in the Fertile Crescent, and more particularly in the mountainous regions that surround the fertile alluvial plains of the Tigris and Euphrates rivers [7]. Lessons from botany, archaeology and more recently from molecular genetics provided more elaborate and precise ideas about the geography and the chronology of the different events, which will be synthesized in the present paper. The case of wheat, however, is far more complex than that of barley or maize since the Triticum genus is made of several species with various ploidy levels, from 2 × (14 chromosomes) to 6 × (42 chromosomes).
The common ancestor of the grass (Poaceae) family, with putatively five chromosome pairs [8], produced a diploid ancestor of the Triticeae subtribe with seven chromosomes. As illustrated in Fig. 1, the first polyploidisation event occurred at 500,000–150,000 years BP (before present), between Triticum urartu (genome AuAu) and an unknown, possibly extinct species closely related to Aegilops speltoides, thus creating a new amphi-tetraploid species with 14 chromosome pairs, namely Triticum turgidum L. ssp diccocoides (or T. diccocoides, genome AuAu BB). This species was then domesticated and evolved as T. turgidum ssp dicoccum Schübl. (or T. dicoccum), which is the progenitor of durum wheat. The second polyploidisation event occurred about 10,000 BP, between the domesticated tetraploid T. dicoccum and the wild diploid species T. tauschii to give the hexaploid species (21 chromosome pairs) Triticum aestivum L. or bread wheat. T. aestivum is only known in its domesticated form, the widely grown bread wheat.
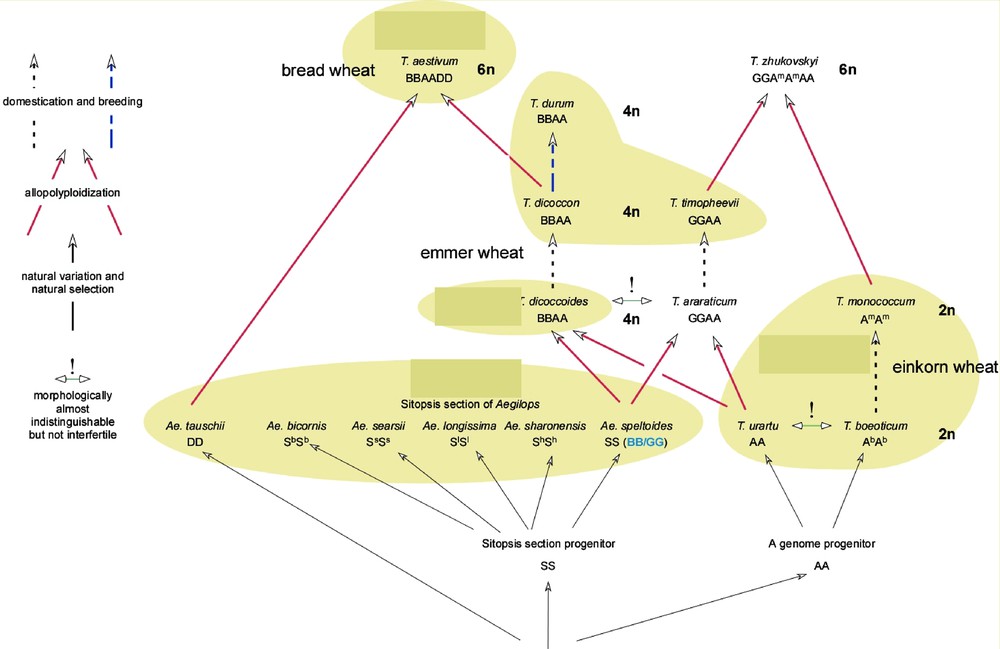
Wheat evolution and events. Wheat (Triticum) and Aegilops species/taxa involved (from Kilian, pers. comm.).
3 Why man turned to farming?
What are the reasons why modern man, Homo sapiens, a hunter and gatherer for 100,000 years, moved to plant cultivation and animal husbandry? Likely we will never know whether the demographic expansion pushed humankind to become farmers to increase food resources, or if the discovery of agriculture, by providing more food, enabled an increase in human population. According to ethnologists, the practice of primitive agriculture required an energy input (kcal) higher than that for hunting and gathering. Agriculture would thus not have been adopted for comfort but rather by necessity. However, this estimate of energy requirement is inferred from data on contemporary hunter-gatherers and might not be appropriate for Neolithic time. Moreover, it seems that the demographic expansion had started with the warming that just followed the last ice age about 13,000 BP, while the oldest remains of T. diccocoides in archeological digs are dated at about 19,000 BP [9]. Yet, the Old World agriculture was probably born in the Fertile Crescent, between 12,000 and 9,500 BP, during the Young Dryas period, characterized in the Middle East by winter frost and rainfall decline (11,000–10,300 BP). This period likely reduced available food resources and led human societies to cultivate the plant species they were used to gather [10]. The debate on the origins of agriculture will probably never end and the verity is probably a mixture of these two hypotheses. Agriculture might have been invented because natural resources were rarefied as a consequence of climate change and perhaps because of demographic pressure. It is not doubtful that a higher soil productivity (perhaps already associated to extended working time) allowed a higher fecundity, leading to demographic expansion, which in turn made a pressure toward higher productivity.
4 The contribution of archaeology
Archaeologists make inferences from the dating of fossilized remains of crops, mostly of grains that are the most easily conserved [11–14]. The main archeological sites are located in the Jordan Valley and the upper valley of the Tigris and Euphrates rivers, as well as in the mountainous regions of South-Eastern Turkey (Fig. 2). However, the archaeological sites that have been discovered may only represent a small and biased sample of the existing Neolithic villages, where domestication might have occurred. Moreover, most ancient sites experienced a long occupancy period, sometimes occurring in several waves during which wild types of wheat, barley, lens and pea grains were gradually replaced by domesticated forms. Morphological differences allow distinguishing brittle (i.e. wild) from non-brittle (i.e. domesticated) rachis, and hulled from free-threshing grains (Fig. 3) However, it is generally difficult to differentiate the free-threshing durum and bread wheat remains.
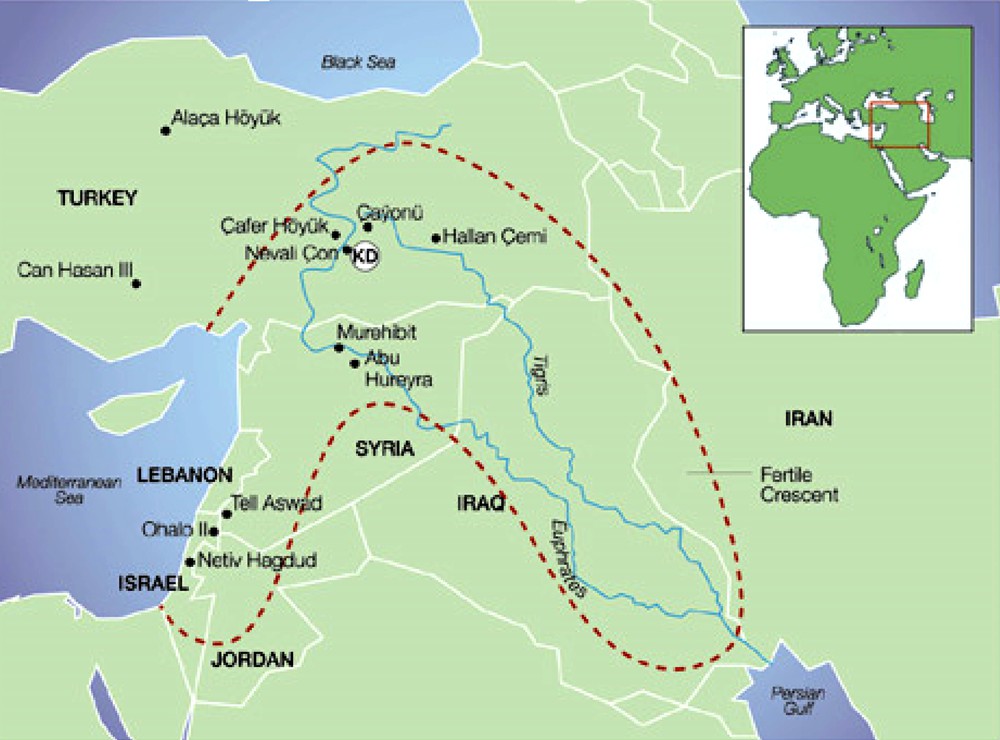
After Salamini et al. 2002: The Fertile Crescent and the main archaeological sites (KD: Karacadag Mountains in Turkey).

A major domestication trait: Brittle rachis of T. diccocoides (a) and non Brittle rachis of T. dicoccum.
For example, on the site of Abu Hureyra in Syria, a majority of wild grains were found for the period from 12,200 to 11,100 BP. Then a progressive substitution by domesticated grains could be observed for the period between 11,000 and 10,500 BP and finally domination by the latter forms is apparent during the period from 10,500 to 7800 BP. The domestication phase thus spread over several centuries and simultaneously concerned several species: einkorn (Triticum monococcum), emmer wheat (T. dicoccum), barley (Hordeum vulgare) and rye (Secale cereale) [13,15]. As a consequence of the limited sampling of sites and the relatively low accuracy of radiocarbon dating, archaeological research can only estimate ranges of dates for domestication in the Fertile Crescent. Genetic research established evidence for locating the cradle of agriculture in the Karacadag Mountains [10,16]. Indeed, most species that were cultivated prior to domestication originated from wild species naturally growing in this region, with the remarkable exception of bread wheat (see later). With the exception of rye, cereals are self-fertilized species. Gene flow via pollen and seed dispersion is limited in nature, which led to a high level of genetic differentiation among local populations. Then, provided that local populations of wild species did not migrate since the Neolithic period, populations most similar to domesticated species are the likely progenitors, thus revealing the primary domestication site(s). Moreover, since population expansion and migration leave imprints in the distribution of fossil DNA, it is possible to trace the migration routes of cultivated plants and to date such migrations [17], as is done for human populations [18].
5 The revolution of molecular genetics
These studies have been carried out during the last two decades since molecular biology has provided a huge number of molecular markers allowing a precise estimation of genetic similarities. For instance, Heun et al. [19] studied 338 accessions of T. monococcum from the Fertile Crescent and other geographic origins from Europe, including 68 domesticated accessions of T. monococcum monococcum, nine weedy accessions of T. monococcum aegilopoides and 194 wild accessions of T. monococcum boeticum. Using 228 Amplified Fragment Length Polymorphisms (AFLP) markers basically located in non-coding sequences and often assumed to be selectively neutral, these authors showed that the wild accessions most related to cultivated accessions originated from the Karacadag Mountains, thereby providing strong evidence for locating the cradle of agriculture in the Middle-East [16]. These results were later on confirmed by Kilian et al. [20], who also demonstrated that the diploid A genome donor of polyploid wheats, T. urartu, was actually genetically distinct from the wild T. monococcum boeticum, the progenitor of domesticated einkorn [21]. Einkorn was likely the first domesticated hulled wheat. It was one of the founder grain crops of Neolithic agriculture in the Near East, and a principal species of the early crop introduction in Europe. However, since the Bronze Age, its importance declined gradually and it is now a relic crop, only being sporadically grown.
Similar studies have been carried out on the wild forms of T. diccocoides and the domesticated forms of the tetraploid wheat T. turgidum [22]. These studies also used AFLP markers and disclosed that the wild populations from Karacadag Mountains were genetically close to old emmer wheats, suggesting a fate very similar to that of einkorn. Alternatively, Mori et al. [23] proposed a diphyletic origin for domesticated emmer, based upon two simultaneous primary domestication events occurring in the Karacadag Mountains and the Kartal region that is located 200 km west from Karacadag. This question was later solved by Ozkan [24] and Luo et al. [25]. By analyzing a larger sample of populations, these authors confirmed their first results and proposed an hypothesis to explain Mori's results in which the diphyletic origin of domesticated emmer would have arisen from a secondary introgression of some domesticated haplotypes from Karacadag into wild populations of Kartal, following their introduction by Neolithic farmers.
Accordingly, a monophyletic origin of both diploid and tetraploid domesticated wheat is now considered as being the most likely hypothesis. We already mentioned that the two A genomes of diploid and tetraploid wheat originated from different wild species. Another wild tetraploid species, T. araraticum, with an AAGG genome composition resembling T. dicoccoides (though they are incompatible) has also been domesticated in the Caucasian region, being the progenitor of T. timopheevi whose cultivation has always been limited. Unlike wheat, cultivated barley (Hordeum vulgare) does seem to have a diphyletic origin. Here, wild populations of H. spontaneum in the western and eastern parts of the Fertile Crescent are the progenitors of European and Asian cultivated barley genotypes, respectively [26,27]. Thus domestication of wild species was at the center of einkorn (T. monococcum) origin, and T. turgidum durum grows today mostly in regions with summer-dry, relatively warm Mediterranean climate. As far as we know, there are no wild species of hexaploid Triticum, which is therefore a new species that evolved during T. dicoccum cultivation, when T. dicoccum crossed with T. tauschii. Molecular genetics provided evidence for this event. Dvorak et al. [28], using molecular markers determined that the D genome of bread wheat originated from the subspecies T. tauschii var strangulata, which naturally grows in Northern Iran and the Trans-Caucasian region. Since T. dicoccoides does not naturally grow in this region, the only conclusion is that the hybridization had only been possible after the domestication of T. dicoccum and subsequent distribution by farmers who swapped agricultural technologies between the Karacadag region, Northern Iran and Transcaucasia, between 8000 and 7000 BP. The first studies based on molecular markers suggested a polyphyletic origin for hexaploid wheat [29,30]. However, the study of the promoter region of the GluDy loci encoding the high molecular weight glutenin subunits (storage proteins) brought evidence for a new diphyletic hypothesis [31]. Indeed these D-genome specific markers support previous studies suggesting the existence of at least two Ae. tauschii sources that contributed germplasm to the D genome of T. aestivum. Ae. tauschii from Syria and Turkey had relatively high nucleotide diversity and possessed all the major GluDy alleles, indicating that these populations are probably ancient and not the result of adventitious spread. The presence in the Turkish population of both of the shared alleles suggests that hexaploid wheat is likely to have originated in Southeastern Turkey or Northern Syria, within the Fertile Crescent and near to the farming villages where archaeological remains of hexaploid wheats were first found. A second and more recent hexaploidisation probably occurred in Northern Iran and Trans-Caucasia, where bread wheat would have appeared about 1300 years later [31]. However, the genotypic diversity of wheat landraces from this region is higher than that found in Turkey, suggesting that Trans-Caucasia is an important centre of secondary diversification. T. tauschii thrives in an area characterized by continental climatic conditions and is a successful colonizer of secondary, man-made habitats. It may have become a common weed in T. dicoccum fields where hybridization occurred. These features of the D genome donor may have contributed to the success of bread wheat and its broad range of adaptation under cultivated conditions. Experimental evidence indicates that the first hexaploid wheat had hulled grains, as for T. aestivum subsp. spelta. Indeed, artificial synthesis of T. aestivum by crossing tetraploid T. turgidum with diploid T. tauschii almost always results in hulled products, irrespective of whether the T. turgidum parent is hulled or naked. It is therefore difficult to infer which form (hulled or naked) hybridized with T. tauschii in Neolithic fields 8000 years ago.
6 Domestication syndrome and consequences of genetic bottlenecks
Man thus started to cultivate wild plant species adapted locally, likely plants of nutrional value possibly corresponding to the species he collected as a hunter and gatherer [4]. How did the traits that characterize the domestication syndrome manifest? By chronologic order, the first of these traits was the non-brittle rachis that has a simple monogenic determinism (Br for brittle rachis [32]). In a wild species, this trait causes a negative fitness due to reduced seed dispersion, progeny and species’ success. Thus a non-brittle rachis mutation that could have appeared in a wild population would have likely been eliminated by natural selection. However, in a cultivated field such a mutation would confer a positive fitness if farmers preferred and nurtured plants carrying this trait. If harvest occurred after brittle-rachis plants had matured and dispersed seed, the seed pool would have been enriched with non-brittle types, which would eventually dominate the population. This seems to have occurred in archaeological sites, as indicated by the progressive replacement of brittle remains by non-brittle rachis types. Population geneticists have estimated the fitness of non-brittle over brittle rachis plants according to various parameters including seed set and seed dormancy, and they used this fitness in demographic models to estimate the time required for total replacement. Such total replacement would have taken 500–1000 years, which fits with the observations from archaeological sites [33,34]. The non-brittle rachis, which is governed by a single gene, could have dominated a population in cultivated fields without conscious selection by Neolithic farmers, simply because this trait would have been advantageous on farms where a portion of the harvested grains is retained as seed for the subsequent growing season, while tilling eliminates part of the seedlings born from brittle rachis plants.
The second trait, called naked grain, presents a slight fitness benefit over hulled grain (Tg, for tenacious glume [32]). This trait is associated with a shorter dormancy, which confers an advantage over the more dormant-hulled grains for germination and growth during optimal conditions. Although naked grains are preferred for their higher quality flour content for making bread, there is no evidence that the first farmer-millers sorted the naked from the hulled grains.
Naked-grain wheat has dominated hulled-grain wheat. However, today the hulled-grain cereals are marginally grown. It is worth noting that a common phenotype, namely non-shattering, can arise from independent mutations in different genes. Yet it was recently shown that the major threshability genes, namely soft glume (sog) in diploid T. monococcum and tenacious glume (Tg) in T. turgidum diccocoides originated from mutations at non-orthologous loci [35]. This case of convergent evolution is not so surprising as we have seen that the A genome of diploid and tetraploid wheat did not derive from the same donor species. Another mutation had a noticeable fate in durum wheat, but especially in bread wheat. Thus, the Q mutation (Q allele) is likely to be at the origin of the extraordinary evolutionary success of this species. This gene has been cloned and found to be homologous to the APETALA 2 gene of Arabidopsis thaliana [36]. The Q allele affects a gene with pleiotropic effects on glume shape and glume tenacity, and on the shape of spike; consequently the spike is more compact with more flowers per spikelet, which increases the number of sinks for assimilate and thus potential yield (Fig. 4). The Q mutation seems to have appeared only once. It has a monophyletic origin. However, it is still unclear whether it appeared first in tetraploid wheat or in bread wheat with a secondary transfer to durum wheat. It seems that the q allele found in T. aestivum spelta with hulled grain appeared about 1000 years later than the first hexaploid wheat with naked grains [36]. Thus the Q allele appears to be the primitive allele in hexaploid wheat, possibly being inherited from a Q-tetraploid wheat that hybridized to T. tauschii. In any case, this mutation played a crucial role in the world distribution of durum and bread wheat. All extant 6 × naked wheat species carry the tg/tg q/q genotype.

A major domestication trait: a) wild (qq) vs b) squared head (QQ). The Q allele affects a gene affecting spike shape of spike.
Events of the domestication syndrome may be considered as completed with the fixation of non-brittle rachis and naked-grain traits in cultivated fields. Then several other traits have progressively evolved in a favorable direction for farmers, namely flowering time and grain size. The mutations responsible for these changes have been fixed and concentrated, where they conferred agricultural benefits. However, when and where the voluntary selection of favorable traits by farmers occurred first is still unclear. Random mutation is an important contributor to phenotypic variation, but most mutations are more likely to be neutral or deleterious than advantageous. It has been postulated that polyploid species like wheat contain higher amounts of genomic DNA, including duplicated genes, thus being more prone to accumulating negative mutations. On the other hand, the duplicated genes will tolerate more mutations and thrive from a process termed subfunctionalization. Therefore, polyploid wheats have more opportunities to wait until a favorable mutation occurs and becomes fixed. This genome plasticity is thought to be at the origin of the evolutionary success of polyploids, in wild as well as in cultivated species [37].
However, the success of wheat domestication is tempered by the degree of genetic variability in tetraploid and hexaploid species compared to diploid and tetraploid wild populations. Indeed, domestication and polyploidisation events likely involved a few plants, which led to a dramatic reduction of effective population size also called genetic bottleneck. The intensity of these bottlenecks and their effects on allelic diversity and distribution has been estimated by different methods based on molecular markers [38,39]. Using coalescent models of sequence evolution, such studies determined that the domestication of T. dicoccum from T. diccocoides had led to a bottleneck associated with a reduction in the effective population size of the domesticated species to about one third of that of the wild populations. However, spontaneous hybridization between domesticated forms and surrounding wild populations may have increased variability and attenuated the bottleneck effect [20]. More recently, even stronger bottlenecks occurred during the selection of modern varieties and therefore estimates of effective population sizes actually measure the effects of these two sources of bottlenecks (domestication then modern selection). This reduction of genetic diversity is even more pronounced for alleles exerting a strong positive selection (e.g., the Q allele) and for genes associated to selective genes by linkage disequilibrium (LD) that generally correspond to closely mapped genes.
Thus, domestication and later modern scientific selection of crop species in an increasingly anthropized environment was globally positive, as documented by the dramatic increase in yield, for example. However, domestication and selection had side effects, such as the loss of some favorable alleles, either because they were in LD with negative alleles at important adaptive genes, or simply by chance.
7 An urgent need to preserve and mobilize untapped genetic resources
With the food crisis of 2007, and perhaps the production losses in Russia and Ukraine noted in 2010, people became aware that food supply might be far from guaranteed. To provide food, feed and raw materials for biobased industries in a world of 9 billion people by 2050, the wheat demand is expected to increase from 621 mt to more than 900 mt [1]. This implies an annual production growth rate of 1.6% from 2005–2020, while it was limited to only 0.9% from 1985 to 1995. Moreover, the rate of yield increase has slowed down from 1995 to 2005 in nearly every country [1,40], being close to 0 in EU particularly in the major producing countries such as France, Germany and UK (Fig. 5). Although genetic progress was continuous at a rate of ∼0.1 t/ha/year, this genetic progress has only been sufficient to compensate the negative effects of abiotic and biotic stresses, but not to allow a further increase in yield [3]. As most scenarios of climate change forecast a global increase of 2–4 °C, with increased variability and frequency of extreme events, the negative effect of climate change on wheat yield is expected to increase [41]. Moreover, there is a general consensus that this production increase should be achieved in sustainable farming systems, i.e. with less use of synthetic fertilizers, pesticides and fossil fuels, which were an integral part of the increased yields after 1930. Plant breeders must respond to these new strategies to develop more efficient varieties, being more productive and less resource-intensive. This unprecedented challenge will require the development of a new knowledge-based science and economy.
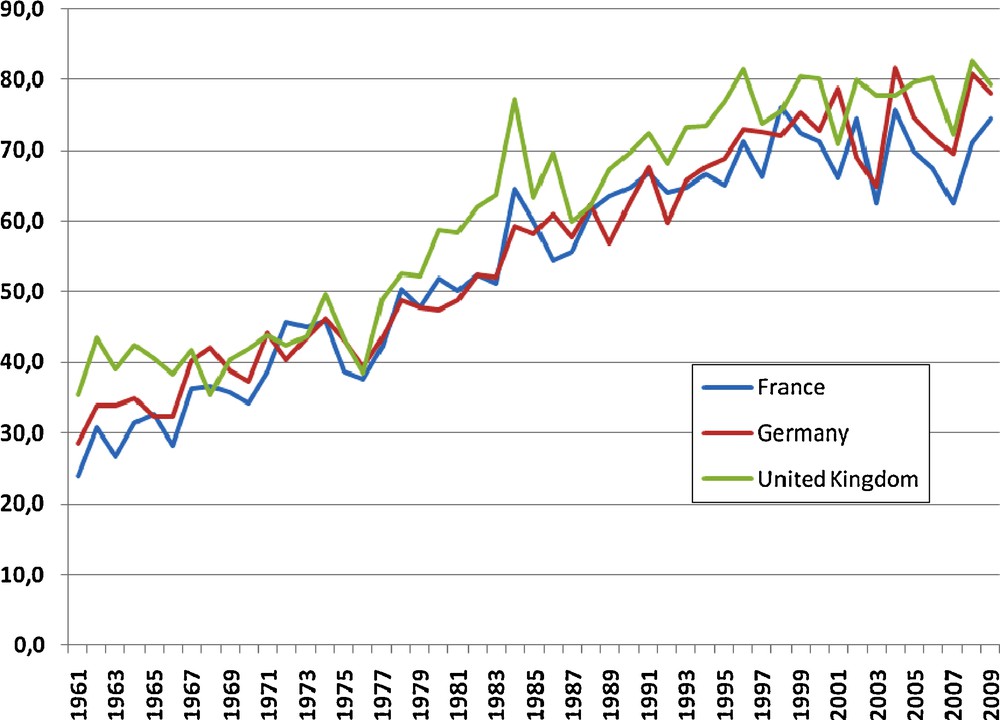
Evolution of average yield (in quintals - 100s kg per hectare) in the three major producing EU countries.
As a consequence of positive selection during domestication and scientific breeding, modern varieties actually contain superior allele combinations for intensive cropping systems, as documented by the continuous yield improvement from 1920 to 1990 (Fig. 5). However, reduced diversity caused by genetic bottlenecks has diminished their potential to further evolve in a changing environment. The frequency of the most adapted alleles has increased, though many other potentially adaptive alleles have been lost [38,39].
Wild crop relatives and landraces are presumed to contain alleles useful for modern breeding programs [42]. Until now, wild wheat relatives have been used to incorporate new sources of resistance to diseases and pests, most being controlled by single genes. However, monogenic resistances are often overcome. A more durable resistance relies on quantitative (polygenic) genetics to sustain resistance against evolving pests. Moreover, traits associated with stress tolerance are probably polygenic and new strategies must be developed to exploit wild relatives for polygenic traits. Therefore, all hitherto neglected sources of allelic variation for genes controlling complex polygenic traits should be revisited. However, identification of useful target genes and alleles in wild ancestor or primitive cultivars suffers from the problem of bias in human perception of important traits. In other words, we tend to investigate what we already think is important, such as disease resistance genes [43]. A newly appeared approach consists in genomic scans comparing diversity at molecular markers in wild vs. domesticated populations. Regions with significant reduction in gene diversity and extension of local LD provide evidence of selection imprinting. This approach gives an unbiased view about the type of genes that have played an important role in domestication, and might be further interesting in breeding for adaptation [44,45]. This illustrates how study of domestication processes at molecular level can help identifying novel target genes for future selection programs.
Usually backcross schemes are used to introgress wild chromosome segments carrying favorable traits into an adapted genetic background [46]. However, this strategy is less suitable for complex multigenic traits although molecular markers can help reducing time and cost of experiments [47,48]. An alternative is to mimic what happened in nature 10,000 years ago, and to give evolution a new chance to play with the allelic richness of wild species and old landraces. Indeed, if domestication occurred only once at a single place, the raw material from which modern varieties evolved only represents a small part of the available range of diversity present in the wild ancestor. Moreover, it is unlikely that this initial bottleneck has sampled the most valuable part of the diversity, but rather did it at random. The idea behind neodomestication is to give a new chance to wild species to give rise to domesticated forms of potential interest in breeding programs. This can be achieved by constructing a multiparental population from a broad genetic base and allowing it to evolve under natural selection pressure [49]. As the rate of allele frequency changes relies on cross-fertilization, it is expected to be rather low in self-fertilized species, and introducing some male-sterility genes may be helpful. However, primary domestication lasted over at least half a millenary, while the urgent need of increasing yield is for the coming 50 years, which represents only 5–10 generation lengths in classical breeding schemes. Neodomestication would have to be much faster. Molecular genetics may provide tools for accelerated selection, which can be applied in the context of neodomestication. Genomic selection [50] consists of replacing phenotypic selection with a marker-based index selection (Genomic estimate of breeding value [GEBV]). Theoretically, genomic selection increases selection intensity and reduces the number of generations for selection. Moreover, dense molecular markers now offer the means to monitor the evolution of genetic diversity across generations of selection, and methods have been developed to balance genetic gain and drift, i.e. avoid diversity losses [51]. Again this is a lesson from the study of domestication and its consequences on crop genetic diversity. The advance of sequencing (www.iwsc.org) will soon provide the high throughput, high density and low cost genotyping tools required for implementing efficient genomic selection, which should also be used in neo-domestication experiments.
However, to make neodomestication possible for future generations, we need to preserve the reservoir of wild species and landraces. This is usually achieved with gene banks but in situ conservation is the best option for wild populations, as they continue to evolve with changing selection pressures. But the adverse effect of the expected climate change is threatening the survival of wild wheat relatives in their natural habitats of the Fertile Crescent. In this region biodiversity indices are pessimistic: the expected increase of drought period duration and severity (aridity index) may not permit wild wheat stands to survive (Fig. 6 [52]). Thus, in situ preservation of wild Triticeae populations at the center of origin should be a priority.
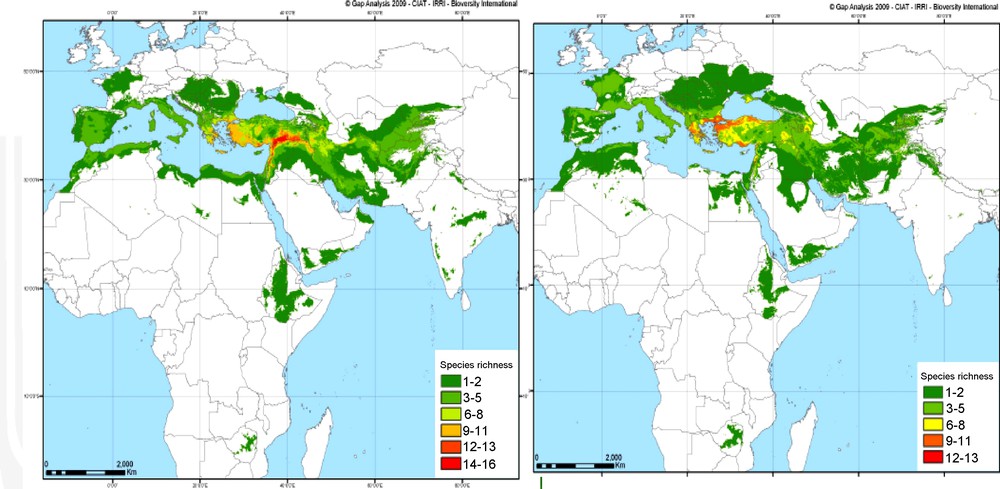
Predicted evolution of wheat-related species richness indices between present (left) and 2050 (right).
Conflict of interest statement
The author has no conflict of interest.
Acknowledgements
The author wishes to thank Dr E. Storlie for thorough revision of the manuscript and his invaluable help in English improvement.


