1 Introduction
Cellulose and hemicellulose are two major components of plant material. Cellulose consists of long polymers of β-1,4-linked glucose units that are organized into higher-order fibrillar structures. Hemicellulose is a group of heteropolysaccharides composed of two or more kinds of monosaccharides such as xylose, arabinose, mannose, glucose, galactose, and glucuronic acid [1,2]. In nature, a large amount of plant biomass is formed annually, making it an attractive potential as a renewable source of energy and basic chemicals [3,4]. Efficient degradation of cellulose and hemicellulose is the bottleneck in fully exploiting this potential [5].
Many microorganisms, mainly filamentous fungi and bacteria, produce a variety of hydrolytic enzymes which synergistically hydrolyze cellulose and hemicellulose to smaller oligosaccharides and finally to corresponding monomers [6,7]. The most studied filamentous fungus in respect to lignocellulose degrading enzyme production is Trichoderma reesei. In T. reesei, most cellulase and hemicellulase genes are coordinately induced by cellulose or some soluble carbon sources (e.g., lactose), and the expression has been shown to be regulated at the transcriptional level [8–10].
In addition to T. reesei, many Penicillium species have been reported with ability of producing high activities of cellulase and hemicellulase and are valuable for industrial application [11–13]. Penicillium decumbens 114-2 isolated in our laboratory is a fast-growing filamentous fungus and secretes a variety of lignocellulolytic enzymes [14]. Its catabolite-repression-resistant mutant JU-A10 with high secretion capacity of cellulolytic enzymes has been isolated [15] and used for industrial cellulase preparation production and cellulosic ethanol research in China [16]. However, less is known about the regulation of cellulase and hemicellulase expression in P. decumbens, which might be different from that in T. reesei. To gain a better understanding of the regulatory mechanisms of lignocellulolytic enzyme synthesis in P. decumbens, and whether this regulation was changed in mutant JU-A10, transcription of several cellulase and hemicellulase genes in wild-type strain 114-2 and JU-A10 was analyzed in this study. According to the result of secretomic analysis of 114-2 grown on the medium with cellulose-wheat bran as carbon sources (unpublished result), 6 major lignocellulolytic enzymes were selected from dozens of lignocellulolytic enzymes for transcription analysis, including 2 endoglucanase genes (cel7B/egl1 and cel5A/egl2), 2 cellobiohydrolase genes (cel7A/chh1 and cel6A/cbh2), and 2 xylanase genes (xyn10A/xyn1 and xyn11A/xyn2). Transcription of these genes in response to 6 different carbon sources (i.e. d-glucose, d-lactose, l-sorbose, d-cellobiose, cellulose, and cellulose-wheat bran) was investigated by real-time quantitative PCR. The results are expected to lead to improvements in controlling cellulase and hemicellulase gene expression and developing strains expressing tailor-made enzyme cocktails needed in industrial applications.
2 Materials and methods
2.1 Strains and culture conditions
The wild-type strain P. decumbens 114-2 and its catabolite-repression-resistant mutant JU-A10 were cultured on wheat bran extract agar (10% wheat bran extract, 2% agar) plates at 30 °C for 4 days, and then the spores were inoculated into the minimal medium for pre-culture. The minimal medium contained (per liter): 3 g KH2PO4, 5 g (NH4)2SO4, 0.6 g MgSO4·7H2O, 0.6 g CaCl2, 7.5 mg FeSO4·7H2O, 2.5 mg MnSO4·H2O, 3.6 mg ZnSO4·7H2O, 3.7 mg CoCl2·6H2O, 1 g peptone and 20 g glucose. After 48 h of cultivation at 30 °C with shaking at 200 rpm, the mycelia were harvested, washed with the same medium containing no glucose and peptone (MNGP), and transferred to MNGP for further cultivation at 30 °C with shaking at 200 rpm.
After 6 h cultivation in MNGP in order for the cells to consume intracellular glucose, 1 mM d-lactose, 1 mM d-cellobiose, 1 mM l-sorbose, 1 mM d-glucose, 1% microcrystalline cellulose, 0.5% microcrystalline cellulose plus 0.5% wheat bran were added into the medium, respectively. The cultures were then incubated at 30 °C with shaking at 200 rpm for 2 h, 4 h or 10 h. The mycelia harvested from culture after cultivation were immediately frozen in liquid nitrogen and stored at −80 °C for mRNA extraction.
2.2 RNA preparation and cDNA synthesis
Samples were ground in liquid nitrogen and about 100 mg of mycelium were used for extraction of RNA. Total RNA was extracted using the RNAiso™ reagent (TaKaRa, Japan) according to the manufacturer's protocol. Total RNA was purified and first-strand cDNA was synthesized using PrimeScript® RT reagent Kit With gDNA Eraser (Perfect Real Time) (TaKaRa, Japan) according to the manufacturer's protocol. Oligo dT primer and random 6 mers were both used for reverse transcription.
2.3 Real-time quantitative PCR
The sequences of the primer sets used for real-time quantitative PCR analysis are shown in Table 1. Accession numbers of cel7B, cel5A, cel7A, cel6A, xyn10A, xyn11A and actin in GenBank are ABY56790, ABY28340, ACV95805, HQ286637, HQ286638, HQ286639 and EU855739, respectively. Real-time quantitative PCR amplification was run on a LightCycler instrument with software Version 4.0 (Roche, Mannheim, Germany). PCR amplification was performed in a total volume of 20 μL containing 7.4 μL water, 0.8 μL of each primer (10 μM), 10 μL SYBR® Premix Ex Taq™ (Perfect Real Time) (TaKaRa, Japan) and 1 μL template cDNA. All real-time quantitative PCR runs were performed in triplicate, and negative controls using a reaction as described above but without addition of reverse transcriptase were performed in order to exclude contamination of genomic DNA. The thermal cycling protocol was as follows: initial denaturation for 2 min at 95 °C followed by 40 cycles of 10 s at 95 °C, 30 s at 61 °C. The fluorescence signal was measured at the end of each extension step at 80 °C. After the amplification, a melting curve analysis with a temperature gradient of 0.1 °C s−1 from 65 °C to 95 °C was performed to confirm that only the specific products were amplified. The standard curve of each gene was constructed according to the method reported by Lee et al. [17]. Based on the data from real-time quantitative PCR, the copy number of gene expression was calculated by comparing to standard curve of each gene, respectively. The transcript number of actin gene was quantified as an internal standard. Data processing and statistical analyses were carried out using the Microsoft Excel software. The average values of the 3 determinations for each gene were considered as its expression level. The standard deviation for each gene's expression level was below 10%.
Primers for real-time quantitative PCR.
| Primer name | Sequence (5′-3′) | First nt positiona | Tm (°C) | Fragment size (nt) |
| act-F | CTCCATCCAGGCCGTTCTG | 1390 | 60.9 | 169 |
| act-R | CATGAGGTAGTCGGTCAAGTCAC | 1559 | 58.9 | |
| cel5A-F | AACGCCGACGCTTTCAA | 1156 | 58.0 | 167 |
| cel5A-R | CACGAGGTCCATCCAAGGTAA | 1437 | 59.8 | |
| cel7B-F | AACCTGGAAGAACGGCACC | 593 | 59.6 | 131 |
| cel7B-R | CCTTGTCACAGTCATCGGAGC | 724 | 60.2 | |
| cel7A-F | GTACTTGCGATCCTGATGGG | 832 | 55.8 | 105 |
| cel7A-R | CCACGGTGAAGGGAGACTTG | 937 | 59.1 | |
| cel6A-F | CTCCACCTGCCCATCTTACAC | 1299 | 59.2 | 158 |
| cel6A-R | CCCAGGCTTGTTGCTTAGTG | 1601 | 59.9 | |
| xyn10A-F | GGTCTCCAGGCTCACTTCATC | 934 | 56.4 | 134 |
| xyn10A-R | GTCGAGGGCAAGTTCATACG | 1068 | 58.2 | |
| xyn11A-F | AGAACTTCGGAAGCTACAACCC | 447 | 60.2 | 156 |
| xyn11A-R | TCTTGCGAATGGACCAGTATTG | 603 | 55.9 |
a Nucleotide numbered according to their distance from the 5′ end of genes (positive strand).
3 Results and discussion
3.1 Effects of different carbon sources on enzyme gene expression in 114-2 and JU-A10
The results shown in Fig. 1 give a general picture of the potential of cellulase and xylanase genes expression in two strains grown on different carbon sources at 2 h. In wild-type strain 114-2, glucose clearly repressed expression of all cellulase and xylanase genes. It is not strange since glucose was generally considered as a repressor of cellulolytic genes in filamentous fungi [18,19]. As the intermediate of cellulose enzymatic hydrolysis, the role of cellobiose (and other cello-oligosaccharides) was considered to be important in the inductive effect of cellulose. Cellobiose was reported to act as an active inducer when fed at a low concentration or β-glucosidase inhibitor was added into the medium [20–22]. In T. reesei, it was supposed that induction by cellobiose depends on the competition of cellobiose permease with extracellular β-glucosidase [23]. The similar induction mechanism may exist in P. decumbens. In our study, 1 mM cellobiose appeared no inductive effect in 114-2 and gene expression levels were similar to those on glucose. Minor glucose (50 mg/L) but no cellobiose was detected in the culture, suggesting that cellobiose was readily hydrolyzed to glucose by extracellular β-glucosidase rather than transported into cells. Besides, expression of xyn10A was notably lower than those of other genes when grown on glucose or cellobiose in 114-2, indicating a more strict repression for this gene.
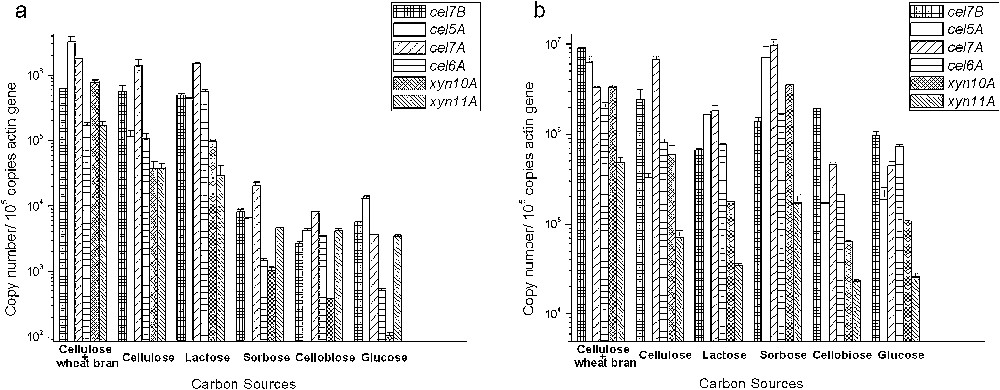
Gene expression of cellulases and xylanases in wild-type strain P. decumbens 114-2 (a) and the mutant JU-A10 (b) in response to 6 different carbon sources at 2 h. Expression of the actin gene was analyzed as a control to compare expression levels between different culture conditions. Copy number was normalized by that of actin transcripts from the same sample.
Cellulose alone or cellulose-wheat bran efficiently induced cellulase and xylanase at transcriptional level. Expressions of all 6 genes tested were significantly increased when cultured on cellulose: transcript numbers of gene cel7B, cel5A, cel7A, cel6A, xyn10A, and xyn11A increased more than 100-, 9-, 380-, 211-, 363-, and 11-fold in 114-2, respectively, compared to that grown on glucose. When cellulose and wheat bran were simultaneously added into the medium, the inductive effect was stronger than cellulose added alone, especially for the 2 xylanases (21- and 5- fold higher than that on cellulose medium for xyn10A and xyn11A, respectively). Soluble cello-oligosaccharides and xylo-oligosaccharides contained in wheat bran might contribute to the enhanced induction [24], and hemicelluloses (mainly arabinoxylans) in wheat bran should be the reason for significantly higher expression of xylanases on this medium. Our previous study showed that either wheat bran or microcrystalline cellulose alone induced much less cellulase and xylanase than their mixture with the same concentration [24], which highlighted the importance of mixed carbon source optimization for overproduction of lignocellulolytic enzymes [11]. Lactose clearly induced expressions of all tested genes with 6- to 1120-fold increase compared with those on glucose or cellobiose medium. Compared with cellulose induced culture, the expression level of gene cel7B, cel7A, xyn10A and xyn11A on lactose medium showed no or very little difference, while the transcription level of gene cel5A and cel6A increased nearly 4-fold and 5-fold, respectively.
Compared to wild-type strain 114-2, similar responses to some carbon sources were observed in mutant JU-A10. Cellulose alone or cellulose-wheat bran efficiently induced all genes tested in mutant JU-A10. The levels of gene expression on cellobiose were also similar to those on glucose. However, gene expressions on lactose and sorbose were different from those in 114-2. For JU-A10, no obvious inductive effect of lactose was detected, and the concentration of lactose was nearly unchanged after 2 h. Since the weak capability of lactose utilization for JU-A10, we suggest that lactose transport or metabolism may have been destructed in this mutant.
Interestingly, l-sorbose was found to induce cellulase and xylanase genes coordinately at the transcriptional level in mutant JU-A10, while no such effect was detected in 114-2. As a monosaccharide, l-sorbose was reported to induce cellulase gene expression in T. reesei [25], but the induction mechanism is totally unclear. We have found that both P. decumbens strains in this study showed very weak growth when cultured on the medium with 1% (w/v) l-sorbose as the sole carbon source (data not shown), indicating the inductive effect in JU-A10 might be not related to its metabolism. On the other hand, the result made it valuable to test whether addition of l-sorbose to the industrial medium would enhance cellulase production by this P. decumbens mutant.
3.2 Difference between cellulose-mediated induction and lactose-mediated induction in wild-type strain
In this study, both soluble lactose and insoluble cellulose significantly induced cellulase and xylanase genes expression, but the inductive effects were somewhat different. In wild-type strain 114-2, high lactose-induced expression of 6 genes was detected at 2 h. Then their expression levels decreased sharply (Fig. 2). At 4 h, the expression levels of cel5A, cel7A and xyn10A decreased down to 1/50, 1/90 and 1/330 respectively with comparison of their expression at 2 h. This decrease might be due to the low concentration of lactose (1 mM) which was consumed rapidly. However, on cellulose, as an insoluble inducer, the expression levels were much steady from 2 h to 10 h. The maximum expression levels of cel5A, cel7A and xyn10A emerged at 4 h, somewhat later than on lactose. The expression levels of cel7B and xyn11A increased even after 4 h. The delay reflects the necessity of formation of a soluble inducer which should be produced by hydrolysis of cellulose.
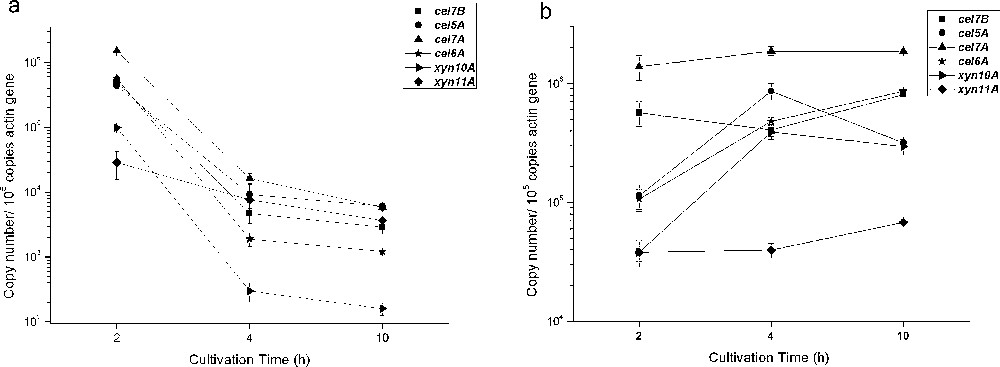
Time course of cellulases and xylanases expression in P. decumbens 114-2 induced by lactose (a) and cellulose (b). Copy number of each gene was normalized as described in Fig. 1.
Lactose is a soluble inducer of cellulase formation and used economically for cellulase production in T. reesei [26]. However, in our study, no inductive effect was found on cellulase production of P. decumbens 114-2 when 20 g/L lactose was used as the sole carbon source (data not shown). The result was consistent with two similar reports, in which no inductive effect was found on cellulase production when lactose (10 g/L and 5 g/L, respectively) was used as carbon source for 2 Penicillium species (P. echinulatum and P. funiculosum) [27,28]. In these studies, too much lactose was added to support the rapid growth of hypha. Catabolite repression caused by glucose might be the reason for repression because lactose was hydrolyzed to d-glucose and β-d-galactose by β-galactosidase [29]. In this study, low concentration of lactose was added to a resting cell system and induced the cellulase gene expression significantly. The result showed 3 speculations: (1) low growth rate of hypha is indispensable for lactose-mediated induction, which is consistent with the result in T. reesei that d-galactose indeed induces cellulases at low growth rate [30]; (2) lactose-mediated induction of cellulase in P. decumbens is concentration-dependent; (3) slow feeding of low concentration lactose during later period of fermentation might improve cellulase and xylanase synthesis.
3.3 Catabolite-repression resistance of mutant strain JU-A10
For all carbon sources tested in this study, the expression level of all 6 genes was substantially higher in JU-A10 than that in wild-type strain 114-2 (Fig. 1). It is worth to note that the folds of increase were much higher in glucose culture than those in cellulose culture. In cellulose culture, the expression levels of cel7B, cel5A, cel7A, cel6A, xyn10A and xyn11A in JU-A10 were just 4-, 3-, 5-, 7-, 15- and 2-fold higher than that in 114-2, respectively (Fig. 3). While in glucose culture, their expression levels in JU-A10 were 170-, 15-, 125-, 1440-, 1030- and 7-fold higher than that in 114-2, respectively (Fig. 3), indicating an obvious derepression of lignocellulolytic enzyme expression in JU-A10.
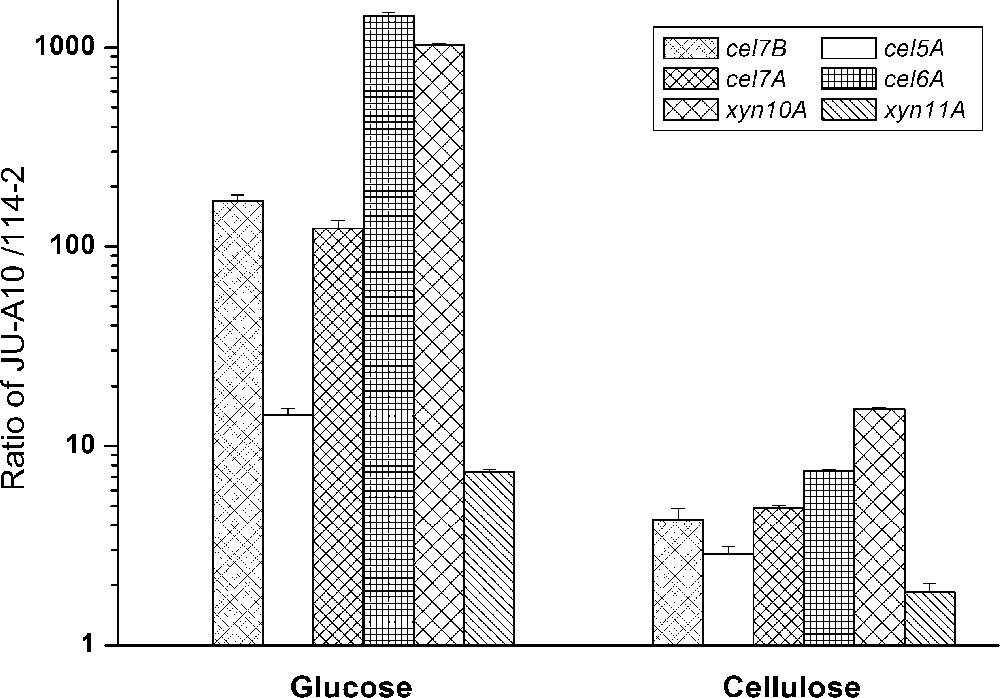
Up-regulation of cellulase and xylanase genes in P. decumbens JU-A10 grown in glucose and cellulose compared with 114-2.
Early studies in some fungi showed creA as the major regulatory gene mediating carbon catabolite repression [31–33]. In T. reesei and Aspergillus spp., deletion of creA or its mutagenesis not only derepressed enzyme gene expression on glucose but also led to a higher expression of enzyme genes even in inducing conditions [10,18,31,32]. Another transcription factor ACEI was also isolated and characterized in T. reesei, and deletion of ace1 led to increased expression of major cellulase and xylanase genes in inducing conditions [34]. Genes encoding homologs of creA and ace1 have been cloned from P. decumbens 114-2, and their roles in expression regulation of lignocellulolytic enzyme genes will be studied in future.
4 Conclusion
In this article, genes encoding 4 major cellulases and 2 major xylanases were proved to be coordinately regulated at transcriptional level. The mixed carbon source cellulose-wheat bran induced the highest expression of most cellulase and xylanase genes in both strains. Lactose at low concentration was first reported to induce the expression of lignocellulolytic enzymes in Penicillium spp. Transcription levels on glucose, lactose and sorbose in mutant JU-A10 were quite different from those in 114-2, suggesting mutations in JU-A10 had changed its responses to these carbon sources roughly. Now a comparative genomic research is in progress to explain the mechanism of improved cellulase production in the mutant. Understanding of expression regulation of lignocellulolytic enzymes in P. decumbens would be helpful for directional cellulase-hyperproducing strain construction.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgments
This work was supported by grants from National Basic Research Program of China (No. 2011CB707403), the National Natural Sciences Foundation of China (No. 31030001), Shandong Provincial Natural Science Foundation, China (ZR2010CM003) and China Postdoctoral Science Foundation (20090461230).


