1 Introduction
Lignocellulose is a complex of carbohydrate polymers (cellulose and hemicellolose) tightly bound to lignin, and is a major constituent of a wide variety of materials including waste materials from agriculture, forestry, wood-based industries, and municipal solid waste [1]. These materials are produced in abundance, and represent a good option for conversion to useful, high value products. Lignocellulose conversion requires a pre-treatment step to degrade or loosen the recalcitrant and heterogeneous lignin fraction. This multi-faceted challenge is being addressed by an ever-increasing suite of ligninolytic enzymes isolated from various sources. Among these, fungal laccases (benzenediol:oxygen oxidoreductase, EC 1.10.3.2) are known to play an important role in lignin degradation/modification processes. These enzymes can be successfully applied to paper manufacturing, enhancement of fibre properties, production of improved forages and pre-treatment of lignocellulosic biomasses for fuel production.
Biotechnology can contribute to the development of “green tools” for the transformation of lignocellulosic feedstocks by providing tailor-made biocatalysts based on the oxidative enzymes responsible for lignin attack in nature [1]. With this purpose, laccases are currently being improved using (rational and random-based) protein engineering [2].
Laccase is one of the oldest enzymes reported and it is arousing great interest in the scientific community because of its very basic requirements (it just needs air to work and its only released by-product is water) and huge catalytic capabilities, making it one of the “greenest” enzymes of the 21st century [3]. This enzyme is produced by various fungi, plants, and certain bacteria or insects [4]. Laccase is able to catalyze direct oxidation of ortho- and para-diphenols, aminophenols, polyphenols, polyamines, and aryl diamines as well as some inorganic ions. It couples the four single-electron oxidations of the reducing substrate to the four electron reductive cleavage of the dioxygen bond, using four Cu atoms distributed against three sites, defined according to their spectroscopic properties. Typical metal content of laccase includes one type-1 (T1) copper, and one type-2 (T2) and two type-3 (T3) copper ions, with T2 and T3 arranged in a trinuclear cluster (TNC) [4].
Fungal laccases exhibit a similar molecular architecture organized in three sequentially arranged cupredoxin-like domains. Each of them has a Greek key β-barrel topology [4]. T1 is located in domain 3, whilst the TNC cluster is embedded between domains 1 and 3 with both domains providing residues for copper coordination. The structure is stabilized by two disulfide bridges between domains 1 and 3 and between domains 1 and 2.
Laccases are commonly classified as low-medium and high redox potential laccases (HRPLs) according to their redox potential at the T1 site ranging from +430 mV in bacterial and plant laccases to +790 mV in some fungal laccases. The latter are by far the most important from a biotechnological point of view [5]. HRPLs are typically secreted by ligninolytic basidiomycetes, the so-called white-rot fungi [3].
The white-rot fungus Pleurotus ostreatus expresses multiple laccase genes encoding isoenzymes with different properties, being the physiological significance of this multiplicity still unknown [6]. Among these, POXA1b, in addition to its high redox potential (+650 mV) [7], is highly stable at high temperature and in the pH interval of 7 to 10 [8]. Thus, this enzyme is a suitable scaffold for directed evolution experiments, since the likelihood of achieving required improvements without affecting its stability is high.
This article reports the optimization of the functional properties of POXA1b laccase expressed in the yeast Saccharomyces cerevisiae [9]. We integrate these results with a structural analysis of some of the generated mutants that allowed us to suggest some of the reasons, at a molecular level, for their enhanced activity.
2 Experimental
2.1 Strains, media, and plasmids
The S. cerevisiae strain used for heterologous expression was W303-1A (MAT ade2-1, his3-11, 15, leu2-3, 112, trp1-1, ura3-1, can1-100). The plasmid used for S. cerevisiae expression was pSAL4 (copper-inducible CUP1 promoter). S. cerevisiae was grown in selective medium (6.7 g L−1 yeast nitrogen base w/o amino acids and ammonium sulfate; 5 g L−1 casaminoacids; 30 mg L−1 adenine; 40 mg L−1 tryptophane; 50 mM succinate buffer pH 5.3; 20 g L−1 glucose).
2.2 Random mutagenesis
Random mutations were introduced with low, medium, and high frequency of mutation, into the POXA1b encoding cDNAs using GeneMorphTM PCR Mutagenesis Kit (Stratagene, La Jolla, CA). EP-PCR was performed with primers POXA1bfw (ATAAAAGCTTGAATTCATGGCGGTTGCATTCG) and POXA1brev (TAAGGATCCAAGCTT TTATAATCATGCTTC).
2.3 Construction of mutant library
The cDNA resulting from EP-PCR on poxa1b cDNA were cloned in pSAL4 expression vector, digested with SmaI and BglII restriction enzymes, by using homologous recombination expression system of S. cerevisiae. Yeast transformation and selection was performed as already reported [10].
2.4 Library screening
Single clones grown on plate were picked and transferred into 96-well plates containing 30 μL of selective medium per well. Plates were incubated at 28 °C, 250 rpm for 24 h. After 24 h, 130 μL of selective medium was added to each well and the plates were incubated at 28 °C, 250 rpm for 24 h. Thirty microliters of each culture was transferred to a new 96-well plate to measure the OD600 value. The plates were then centrifuged for 10 min at 1500 g, 4 °C, and a suitable volume of supernatant was transferred to a new 96-well plate to perform laccase assay. Phenol oxidase activity was assayed at 25 °C using 2 mM 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS) in 0.1 M sodium citrate buffer, pH 3.0. Oxidation of ABTS was followed by absorbance increase at 420 nm (ɛ 36,000 M−1 cm−1), using Benchmark Plus microplate spectrophotometer (BioRad, Hercules, CA). Enzyme activity was expressed in international units (U). Cultures in shaken flasks were also performed. Pre-cultures (10 mL) were grown on selective medium at 28 °C on a rotary shaker (150 rpm). A volume of suspension sufficient to reach a final OD600 value of 0.5 was then used to inoculate 250 mL Erlenmeyer flasks containing 50 mL of selective medium and cells were then grown on a rotary shaker. Optical density and laccase activity determination were daily assayed.
2.5 Screening of library for stability
The collection of 3300 mutants obtained by random mutagenesis of POXA1b laccase was analysed in three different screenings. First and second screening were effectuated in 96-well plate, while the third screening was effectuated in shaken flask. In the first screening the library was analysed after one-day growth in 96-well plate. The supernatant was incubated for 48 hours at pH 3 in Robinson and Britton buffer, and then activity towards ABTS assayed. The positive clones were further analysed during a three days growth in 96-well plate. Cellular density and laccase activity production were followed for three days. Every day the supernatant was incubated for 96 hours at 37 °C at pH 3 in Robinson and Britton buffer. Laccase activity was analysed every 24 hours, in this phase only clones that showed greater stability if compared to the wild-type enzyme at pH 3 were selected.
2.6 Molecular modeling
The structure of POXA1b was obtained by homology modeling from the crystal structure of Trametes versicolor (1GYC pdb entry), with which it shares 60% sequence identity. The last 16 residues of POXA1b were modeled using the coordinates of the corresponding residues at the C-terminus of the crystal structure of the Melanocarpus albomyces laccase (1GWO pdb entry). 3D model and in silico mutants were generated using the SWISS-MODEL web server by means of the project mode option that allow to select the template and control the gap placement in the alignment. Refinement of the models has been performed by molecular dynamics simulations. Simulations on the wild-type POXA1b and on the in silico generated mutants 1M9B and 3M7 C were performed with the GROMACS package as already described [10].
2.7 Stability assays
Stability at pH values was measured using citrate-phosphate buffer adjusted at pH 3, 5 and 7.0, and Tris–HCl buffer adjusted at pH 10.
2.8 r4 cDNA construction
To obtain the laccase r4 cDNA the 3′ terminal portion (from the nucleotide 453) of the cDNA coding for 3M7 C was ligated to pSAL4 vector containing the 5′ terminal portion (the first 5′ terminal 452 nucleotides) of 1M10B encoding cDNA, after KpnI digestion of pSAL4 vectors containing the two cDNAs.
2.9 DNA sequencing
Sequencing by dideoxy chain-termination method was performed by the Primm Sequencing Service (Naples, Italy) using specific oligonucleotide primers.
2.10 Decolourization experiments
Batch decolourization experiments have been performed incubating (1 mL final volume of reaction) crude preparation of laccase containing different enzyme amounts (0.1 U, 1 U and 3 U) in three wastewater models: Acid (0.1 g L−1 Acid blue 62, 0.1 g L−1 Acid Yellow 49, 0.1 g L−1 Acid Red 266, 2 g L−1 Na2SO4, pH 5), Direct (1 g L−1 Direct blue 71, 1 g L−1 Direct Yellow 106, 1 g L−1 Direct Red 80, 5 g L−1 NaCl, pH 9) and Reactive (1.25 g L−1 Reactive blue 222, 1.25 g L−1 Reactive Yellow 145, 1.25 g L−1 Reactive Red 195, 1–25 g L−1 Reactive Black 5, 70 g L−1 Na2SO4, pH 10) [11].
Performances of selected laccases in model wastewater decolourization were evaluated by recording light absorption spectra between 280 and 800 nm at different times (10 min, 20 min, 1 h, 2 h, 3 h and 24 h), and comparing them with the corresponding spectra of controls (the waste incubated with the supernatant of yeast cultures transformed with the empty expression vector). Decolourization was calculated as the extent of decrease of spectrum area recorded between 380 and 740 nm with respect to a control sample. All spectra were recorded after 1:100 dilution of the sample in milliQ water. All experiments were carried out in duplicates, and the mean values were taken. The standard deviation for the experiments was less than ± 5%.
3 Results and discussion
The white-rot fungus P. ostreatus expresses multiple laccase genes encoding isoenzymes with different properties, being the physiological significance of this multiplicity still unknown [6]. Investigation of the recently released P. ostreatus genome (http://www.jgi.doe.gov/sequencing/why/50009.html) has disclosed a complex multicopper oxidase family of up to twelve members. The availability of established recombinant expression systems for P. ostreatus POXA1b [9] has allowed the improvement of enzymes features through a combination of rational and random mutagenesis [10,12,13]. Our starting point was the high redox potential laccase POXA1b [14], which exhibits remarkable stability at alkaline pH [8]. The idea behind the evolution strategy is to create an ideal biocatalyst, able to oxidise a wide assortment of substrates, and stable in a broad range of pH. Thus, different screening criteria were applied to search for such catalysts.
3.1 First generation
A library of almost 1100 mutants with low, medium and high range of mutations was generated by error-prone PCR (EP-PCR) using poxa1b cDNA as template [10]. Screening this library for variants with improved activity at pH 3 using the non-phenolic substrate ABTS has allowed the selection of one mutant, named 1M9B. It shows a single mutation (L112F) leading to an improvement of activity but a decrease of stability with respect to the wild-type enzyme (POXA1b) in all the analyzed conditions. In position 112, there is a generally conserved leucine in all laccases from basidiomycetes, although a phenylalanine seems to be conserved in laccase sequences from ascomycetes. POXA1b 3D model shows that the residue 112 is located in the channel through which the solvent has access to the oxygen-reducing T2/T3 site. To elucidate the role played by this mutation, Molecular Dynamic (MD) simulations were performed on the model of the mutant and compared with those of POXA1b. The analyses show a movement of the subdomain around position 112 as a consequence of a conformational rearrangement due to the presence of the bulkier residue of phenylalanine. A significant effect generated by the mutation is observed in the permeability to water of the T2/T3 channel. Residue F112 is located at the entrance of the channel and its steric hindrance affects the passage of water molecules toward the TNC. As a fact, a larger number of water molecules in the T2/T3 channel has been observed for 1M9B. These data could suggest an increased affinity of this mutant toward oxygen molecules, thus justifying its improved specific activity.
3.2 Second generation
1m9b cDNA was used as template for a second round of EP-PCR at low and medium frequency of mutation [10]. A second generation library of 1200 clones was obtained and screened using the same criterium described before. Three mutants, 1L2B, 1M10B (L112F, K37Q, K51N), and 3M7C (L112F, P494T), were selected showing an activity increase of up to three fold with respect to POXA1b.
Concerning 1M10B mutations, it finds out how directed evolution can get the same result of natural evolution, preserving the properties of mutated amino acids. As a fact, positions 37 and 51 are generally occupied by amidic residues. The mutant 3M7C displays a high activity and an up to two fold increased stability at acidic and neutral pH, as well as at high temperature. 1M10B variant is more stable at alkaline pH (about two fold), whereas its stability is almost equivalent to that of POXA1b in the other tested conditions. The mutation P494T is located in the C-terminal loop that has already been ascertained to affect the function of fungal laccases [4]. MD simulations of this mutant and comparison with the wild-type POXA1b revealed a lower flexibility of the subdomain around position 112 probably responsible of its higher stability. On the other hand, an increased mobility of loops forming the reducing substrate binding site, has been observed in 3M7C leading to higher accessibility of water molecules to the T1 copper site, and to an increased activity of the enzyme.
First and second generation libraries (2300 clones) were then screened for variants with improved activity at pH 5 using either the non-phenolic substrate ABTS, and the phenolic one 2,6-dimethoxyphenol (DMP).
When DMP was used as substrate, two new variants (2L4A and 3L7H) endowed with higher enzyme activity (about three fold increase) than the wild-type laccase were selected [12]. Both mutants doubled the stability of the wild-type enzyme at pH 5. Q272H mutation found in 2L4A may stabilize the protein structure at pH 5 allowing additional interactions – electrostatic and hydrogen bonds – between the positively charged imidazolic ring of His272 and the side chain of Asp287.
After screening with ABTS, one clone, 1L9A, was selected, showing an increase of about three fold with respect to wild-type. Besides the parental mutation (L112F), 1L9A also presents the mutation R284H, located in the loop Gly282-Thr289. This loop may play an important role in protein stability [12]. As a fact, concerning its properties, the mutant increases stability at pH 5 (1.5 fold), while loosing the high characteristic stability of POXA1b at alkaline pH.
3.3 Rational design
During the engineering of POXA1b, some of the beneficial mutations discovered in the early stages of evolution were merged in the rational designed R4 mutant. Synthesis of a laccase joining mutations of 3M7C and 1M10B variants [10] was performed to combine the increased stability of 1M10B at alkaline pH and the improved catalytic efficiency of 3M7C [13]. Joining these mutations a two-fold increase in laccase activity with respect to wild-type enzyme was obtained. The main improvement due to the chimer construction is a slight increase in stability at high temperature, and even more at neutral (about four-fold) and alkaline pH values (about two-fold).
3.4 Third generation
The increased stability of R4 makes it an appropriate scaffold to carry out directed evolution [15]. In fact, more stable enzymes should also be more susceptible to evolution since they have higher ability to tolerate functionally beneficial but destabilizing mutations. Therefore, directed evolution of R4 was chosen as strategy to improve its performances [13]. A library of 1000 clones with low, medium and high average of mutation frequency was obtained through EP-PCR. When this new collection was screened by assaying activity towards ABTS at pH 3, two mutants, 4M10G and 1H6C, with higher activity (about four-fold increase) than that of POXA1b wild-type were selected. Both mutants display higher stability than POXA1b at pH 5 (almost four-fold). 1H6C also retains R4 stability features at pH 10 and at pH 7. Sequence analyses of the selected mutants led to the identification of the mutations V126I for the 4M10G variant and V148L for the 1H6C mutant, besides those of the parental R4 enzyme. Position 148 is located in a closely packed region of the domain 2 [16] adjacent to the reducing substrate binding site. The substitution of V with the larger L causes a close contact of the leucine side-chain with the aromatic ring of Y208, thus changing the conformation of the loop 204–208 forming the bottom of the reducing substrate binding site [16]. This change could, in turn, influence the oxidation rate of the reducing substrate but also the interaction between the domains 2 and 3 [13].
Libraries generated after three rounds of molecular evolution (3300 clones) were then screened for variants with improved activity at pH 8 using the phenolic substrate 2,6 DMP. One variant, 1L10A, was selected for its higher activity (more than four-fold increase) at pH 8 respect to wild-type POXA1b. This mutant loses wild-type stability at pH 7, while raises stability at pH 5 of three fold.
To complete the panel of laccase-based biocatalysts, new selection rounds have been performed on POXA1b mutants libraries. This step has been aimed to develop new laccases more stable than the wild-type at pH 3. One mutant, 3L2A, with higher stability (up to three fold increase) was selected. This selected variant shows an increased stability at neutral and alkaline pH respect to wild-type, besides the higher stability at pH 3.
3.5 Application of improved laccases
Laccase improvement, along with elucidation of their structure-activity-stability relationships, allows to design and develop more suitable systems for specific industrial applications either in the bioremediation field or for lignocellulose conversion, increasing their real exploitation. An ideal catalyst should be stable and active in extreme conditions of pH, normally found in the industrial wastewaters, as well as able to operate on a wide range of substrates. Here we describe, as an example, the application of mutated laccases to the decolourization of industrial wastewaters. In these experiments, models of acid, direct and reactive dye wastewaters from textile industry – defined on the basis of discharged amounts, economic relevance and representativeness of chemical structures of the contained dyes [11] – were used. POXA1b variants represent good candidates thanks to their properties best suited for wastewater conditions. Seven out twelve of selected POXA1b variants show an increased decolourization ability with respect to wild-type toward the acid model. As a fact, a two fold increase in decolourization percentage respect to that of POXA1b has been obtained. In particular the mutant 3M7C shows a higher decolourization capacity (up to 40% decolourization after 3 hours of incubation). No significant decolourization has been obtained towards the others analyzed wastewaters, such as reactive and direct models. All the trials have been performed in a period of time compatible with the stability of the protein, being POXA1b and its derived mutants very stable in a wide range of pH [10,12,13].
4 Conclusion
Although some laccases are being employed successfully in industry, no natural laccase combines the desired attributes of being stable and active over a wide range of temperatures and pH values with a high reduction potential [17]. High-potential laccases represent a starting point to create an ideal catalyst, endowed with all these desirable characteristics, through directed evolution experiments.
In this research, three generations of libraries have been screened using different criteria, and twelve variants have been selected for their improved features (Table 1). Although no a priori selection criterium for “more stable laccase” has been applied, some mutations with beneficial impacts on total enzyme activity also significantly contributed to protein stability in different environmental conditions. This work represents an example of how random and rational approaches can be combined for the engineering of protein function, and how such a strategy could provide an inventory (Fig. 1) of enzymes better-suited for different industrial applications.
Properties of selected POXA1b variants.
| Mutants | Substitution | Activity | Stability | ||||
| pH 3 | pH 5 | pH 7 | pH 10 | 60 °C | |||
| 1M9B | L112F | 1.5× | – | – | – | – | – |
| 1L2B | L112F, N248Y, N261K, V350I | 2.5× | = | + | – | = | = |
| 1M10B | K37Q, K51N, L112F | 2.5× | = | + | = | ++ | = |
| 3M7C | L112F, P494T | 3× | + | ++ | ++ | = | + |
| 2L4A | L112F, Q272H | 2.7× | – | ++ | = | = | = |
| 3L7H | L112F, S285N, N328S | 2.7× | – | ++ | – | – | = |
| 1L9A | L112F, R284H | 3× | – | + | = | – | = |
| R4 | K37Q, K51N, L112F, P494T | 2.5× | – | = | +++ | ++ | + |
| 1H6C | K37Q, K51N, L112F, P494T, V148L | 4.5× | – | +++ | +++ | ++ | + |
| 4M10G | K37Q, K51N, L112F, P494T, V126I | 4.5× | – | ++ | ++ | + | = |
| 1L10A | K37Q, V48I, K51N,A391T | 4.5× | – | +++ | – | + | = |
| 3L2A | K37Q, K51N, L112F, S285N | 3×a | ++ | = | +++ | ++ | = |
a This value represents an increase of stability at pH 3 with respect to the wild-type enzyme.
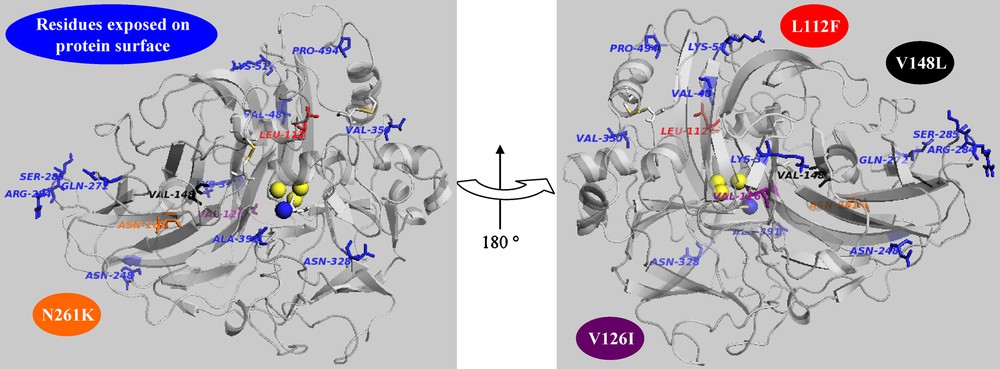
Ribbon representation of POXA1b model. The mutated residues are in licorice rendering. Trinuclear coppers and T1 copper are highlighted in van der Waals representation (yellow and blue, respectively).
Though recombinant production yield of P. ostreatus native and mutated laccases in the yeast S. cerevisiae achieves modest levels (0.2–1 mg l−1), the developed system represents an invaluable tool for rapid screening and selection of new improved laccase variants. Then, the development of new heterologous expression systems in optimised hosts, i.e. filamentous fungi, will provide higher enzyme yields ensuring the exploitation of the selected “tailored” laccase mutant(s) for biotechnological applications on large-scale.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgments
This work is supported by grants from the Ministero dell’Università e della Ricerca Scientifica (Progetti di Rilevante Interesse Nazionale, PRIN), from the Ministero Degli Affari Esteri di Intesa con il Ministero dell’Università e della Ricerca (Progetti di ricerca di base e tecnologica approvati nei protocolli di cooperazione scientifica e tecnologica bilaterale come previsto dal protocollo bilaterale tra Italia e Turchia), from Compagnia di San Paolo, Turin-Italy, project “Sviluppo di procedure di biorisanamento di reflui industriali (BIOFORM),” and from COST Action FP0602 “Biotechnology for lignocellulose biorefineries (BIOBIO)”.


