1 Introduction
Metallothioneins (MTs) are metal-containing proteins that bind zinc, copper and other metallic ligands thanks to the cysteine residues of their polypeptide chain [1]. In addition to supporting homeostatic buffering of metallic micronutrients into the cells by providing Zn and Cu supply, MTs fulfil a broad range of other functions, including detoxification of toxic metals such as cadmium and mercury, scavenging of harmful reactive oxygen species, and others [2,3]. Thus it is not surprising that the versatile MTs are found from bacteria to fungi, protists, plants, and most animal groups [4,5].
In spite of many studies on MTs, little is known about their origin, evolution and diversification [6].
MT evolution is still an obscure aspect of these fascinating metal-binding proteins. The members of the MT protein superfamily have evolved through rounds of duplication and loss events leading to the current heterogeneous scenario. Difficulties in drawing straight evolutionary relationships could be in part attributed to the lack of detailed comparative studies.
Among vertebrates, a great deal of knowledge on MT structure and function relies on studies carried out on mammalian MTs. In mammals, four tandemly clustered genes (MT1 to MT4) are known. Although all genes encode for conserved peptide chains retaining 20 invariant metal-binding cysteines, MT3 and MT4 have developed additional properties relatively to MT1 and MT2, such as protection against brain injuries [7,8] and epithelial differentiation [9], respectively. In addition, the evolution of the MT1 gene in humans led to duplication events that resulted in more than 10 duplicate isoforms [10,11].
Regarding non-mammalian vertebrates, in the last past years many data were collected on the molecular, biochemical and phylogenetic characteristics of piscine and avian metallothioneins [12–18]. Recently, we performed a comparative analysis integrating phylogenetics, functional, and structural approaches, that unravelled the complex evolutionary pattern of the reptilian MT, characterized by gene duplication and gene loss events [19,20]. These data also disclosed for the vertebrate MTs an unexpected complexity in the primary structure that might suppose some functional differences in proteins.
At the moment, however, still very few data are available on the structure of the amphibian metallothioneins [21–23].
To provide a more up-to-date view of the MT genes in vertebrates, we decided to increase the number of the available amphibian MT primary sequences by using a molecular approach. Then, we derived a new vertebrate gene phylogeny taking into account also this animal group.
2 Materials and methods
2.1 Metallothionein cDNA cloning and sequencing
For this study, we cloned and sequenced the MT cDNA from the Italian crested newt Triturus carniflex (Amphibia, Urodela) and the frog Rana esculenta (Amphibia, Anura). Samples for both animals were caught in the outskirt of Naples, under the permission of the Italian Health Ministry.
Liver slices (50–100 mg) were submerged in RNAlater RNA Stabilisation Solution (Ambion) immediately after collection and stored at −20 °C until used. Tissues were homogenized in a small glass homogenizer and the total RNA was purified using the TRI-REAGENT solution (Sigma-Aldrich). The integrity of the total RNA was checked in 1% denaturing formaldehyde agarose gel. Reverse transcription (RT) was carried out using SuperScript II reverse transcriptase according to the standard protocol described by the manufacturer (Invitrogen). The RT reaction was performed at 42 °C for 60 min using the oligo(dT)-adaptor primer previously described [24]. PCR reactions were carried out on 2 μl of first-strand cDNA, using a non-specific primer complementary to the oligo(dT) adaptor and the specific 18mer Amex-Nter (ATGGACTGCGCATGCGCCACT) designed on the basis of the first 7 amino acid residues of Ambystoma mexicanum metallothionein [23]. The purified PCR products were directly cloned into the pCRII-TOPO vector and transformed into TOP10 E. coli cells using the TOPO-TA cloning kit according to the manufacturer's instructions (Invitrogen). Several randomly selected colonies were inoculated into LB broth containing 100 μg/ml ampicillin and incubated overnight at 37 °C with shaking. Plasmids containing inserted sequences were purified using Fast Plasmid columns (Eppendorf) and sequenced using automated methods on an ABI PRISM Genetic Analyzer (PE Biosystems). Sequences and annotations of these two MT cDNAs appear in the DDBJ/EMBL/GenBank database with accession numbers shown in Table 1.
Organism, gene, accession number and acronym of the 41 sequences used for the phylogenetic analysis of the vertebrate metallothioneins.
| Organism | Gene name | Accession No. | Acronym |
| Mammals | |||
| Homo sapiens | Metallothionein 1 | P04731 | H.sapMT1 |
| Metallothionein 2 | P02795 | H.sapMT2 | |
| Metallothionein 3 | P25713 | H.sapMT3 | |
| Metallothionein 4 | NP_116324 | H.sapMT4 | |
| Bos taurus | Metallothionein 1 | P67983 | B.tauMT1 |
| Metallothionein 2 | P68301 | B.tauMT2 | |
| Metallothionein 3 | P37359 | B.tauMT3 | |
| Metallothionein 4 | Q05B43 | B.tauMT4 | |
| Rattus norvegicus | Metallothionein 1 | P02803 | R.norMT1 |
| Metallothionein 2 | P04355 | R.norMT2 | |
| Metallothionein 3 | P37361 | R.norMT3 | |
| Metallothionein 4 | D3ZTJ2 | R.norMT4 | |
| Canis lupus | Metallothionein 1 | O19000 | C.lupMT1 |
| Metallothionein 2 | Q9XST5 | C.lupMT2 | |
| Metallothionein 3 | AB001388 | C.lupMT3 | |
| Metallothionein 4 | BAA87326 | C.lupMT4 | |
| Birds | |||
| Phalacrocorax carbo | Metallothionein 1 | A4PBS7 | P.carMT1 |
| Metallothionein 2 | A4PBS8 | P.carMT2 | |
| Anas platyrhynchos | Metallothionein 1 | A4PBS9 | C.livMT1 |
| Metallothionein 2 | P68494 | C.livMT2 | |
| Gallus gallus | Metallothionein 1 | A4PBT0 | G.galMT1 |
| Metallothionein 2 | P68497 | G.galMT2 | |
| Reptiles | |||
| Anguis fragilis | Metallothionein | AM087390 | A.fraMT |
| Chalcides chalcides | Metallothionein | AM087392 | C.chaMT |
| Phelsuma barbouri | Metallothionein | AM087397 | P.barMT |
| Podarcis sicula | Metallothionein | AJ609541 | P.sicMT |
| Elaphe quatorlineata | Metallothionein | AM087393 | E.quaMT |
| Furcifer pardalis | |||
| Metallothionein | AM087394 | F.parMT | |
| Amphibians | |||
| Ambystoma mexicanum | Metallothionein | AF008583 | A.mexMT |
| Xenopus laevis | Metallothionein | Q05890 | X.laeMT |
| Xenopus tropicalis | Metallothionein | NP_001165150 | X.troMT |
| Rana esculenta | Metallothionein | HE681912 | R.escMT |
| Triturus carniflex | Metallothionein | HE681911 | T.carMT |
| Osteichthyes | |||
| Chionodraco hamatus | Metallothionein I | Y12580 | C.hamMT1 |
| Metallothionein II | Y12581 | C.hamMT2 | |
| Trematomus bernacchii | Metallothionein I | AJ011585 | T.berMT1 |
| Metallothionein II | Z72485 | T.berMT2 | |
| Gymnodraco acuticeps | Metallothionein I | AJ007560 | G.acuMT1 |
| Metallothionein II | AJ007561 | G.acuMT2 | |
| Danio rerio | Metallothionein I | NM131075 | D.rerMT1 |
| Metallothionein II | NM194273 | D.rerMT2 |
2.2 Multiple alignments and sequences analyses
Sequences of vertebrate MTs were retrieved from the NCBI database. The accession numbers of the selected sequences are reported in Table 1. Multiple alignments of MT amino acid sequences were obtained using the program MUSCLE version 3.8 [25]; the amphibian MT coding sequences plus the 3′-UTRs were aligned by the same program. All the alignments used for the phylogenetic analyses were refined manually by the program Se-Al v. 20a11.
The average hydropathic index for each protein sequence was calculated as previously described [15].
2.3 Phylogenetic analyses
Phylogenetic trees were inferred by Maximum Likelihood (ML) model. Analyses were performed with SeaView version 4 software [26], using the Dayhoff + I + G (Invariant sites and Gamma-distributed rates for sites) substitution model following the results from ProtTest for the amino acid sequences [27] and the Generalised Time Reversible (GTR) model following the results from ModelTest for the amphibian MT nucleotide sequences [28]. Bootstrap branch support was estimated using 1000 data sets. Trees visualization and final edition were performed in FigTree v1.3.1.
3 Results
3.1 Characteristics of amphibian metallothioneins
In order to increase the number of known primary structures of the amphibian MT, we cloned and sequenced the cDNA encoding the MT from two species belonging to the class Amphibia: the Salamandrinidae T. carniflex and the Raninae R. esculenta. The MT cDNA fragment obtained by PCR from the hepatic total RNA of the urodelian T. carniflex contains an open reading frame of 189 bp plus a 3′-UTR made of 155 bp; the predicted amino acid sequence corresponds to a protein of 63 amino acids, 20 of them being cysteines. The R. esculenta MT cDNA fragment is of 308 bp, with a coding sequence of 186 bp corresponding to a protein of 62 amino acids with 20 cysteines.
The multiple alignment of these two new nucleotide sequences with the other amphibian MT sequences available is reported in the Fig. 1. Noteworthy, the alignment shows the presence of gaps in the coding sequence region and marked differences in the length of the 3′-UTR sequences, that vary from 641 bp for the A. mexicanum sequence to 122 bp for the R. esculenta MT.

Alignment of the five available amphibian MT nucleotide sequences. Grey box indicates termination codons; asterisks indicate identity among sequences. The GenBank accession numbers of these sequences are reported in Table 1.
The alignment of the deduced amino acid sequences of the amphibian MTs although reveals a good degree of overall similarity among sequences (about 70%), at the same time shows significant differences (Fig. 2). Anuran MTs share the same number of amino acid residues (62), however only Xenopus MT is characterized by the presence of a histidine residue, typical of avian MTs. The two urodelian MTs show an unexpected diversity between each other: T. carniflex MT is made of 63 amino acids, A. mexicanum MT of only 60 amino acids, as piscine MTs.

Alignment of the amphibian MT proteins deduced from the nucleotide sequences reported in Fig. 1.
3.2 Phylogeny of amphibian metallothioneins
The phylogeny of MTs from amphibians was reconstructed with the maximum likelihood method. A phylogenetic tree was constructed using as dataset the amino acidic sequences, and another tree was constructed using as dataset the nucleotide sequences, including the 3′-UTRs. The 3′-non coding regions are highly variable, thus carry more phylogenetic information with respect to MT coding sequences that have a low phylogenetic resolution, due to their relatively short length and low variability. Both the inferred trees show the same topology (Fig. 3), supported by high values of bootstrap analysis. This topology clearly shows a marked incongruence with the phylogeny of extant Amphibia [29].
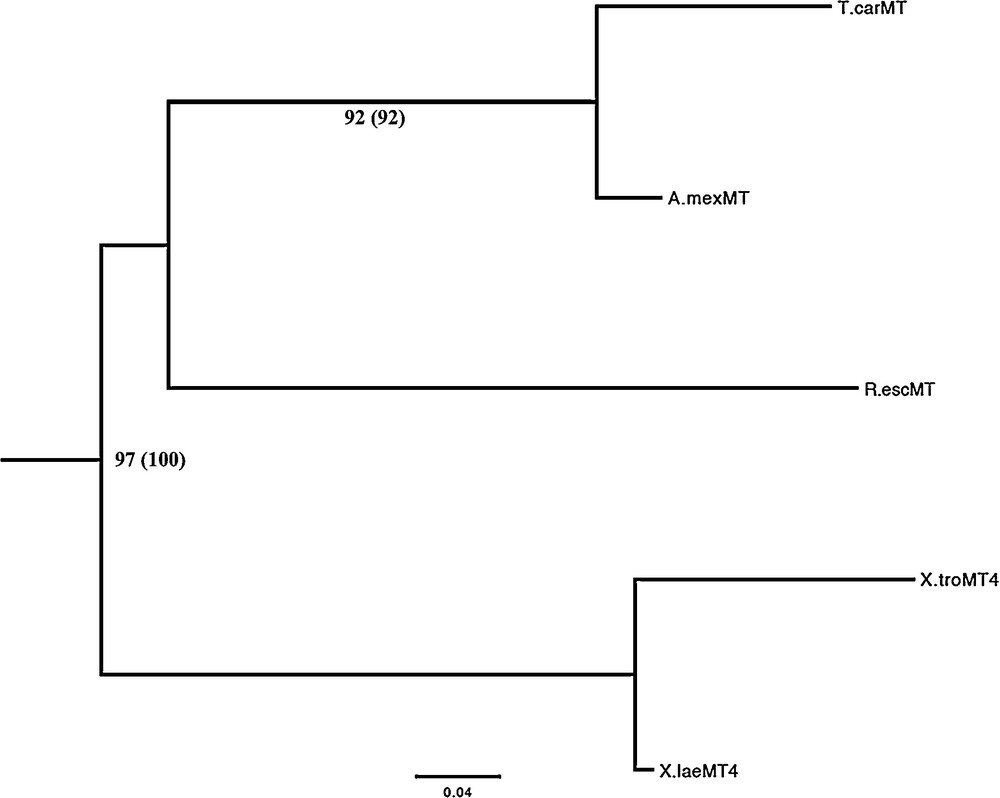
Phylogenetic tree of the amino acid sequences of the amphibian MT inferred by the Maximum Likelihood method using the Dayhoff + I + G substitution model implemented in the software SeaView v. 4.0. The same topology was obtained by using as dataset the nucleotide sequences (coding sequences plus the 3′-UTRs) of the amphibian MT object of the present study and the GTR as substitution model. The numbers at the branches represent the bootstrap values (1000 replicates); in parentheses bootstrap values obtained using the nucleotide sequences. Masquer
Phylogenetic tree of the amino acid sequences of the amphibian MT inferred by the Maximum Likelihood method using the Dayhoff + I + G substitution model implemented in the software SeaView v. 4.0. The same topology was obtained by using as dataset the nucleotide sequences ... Lire la suite
3.3 Phylogeny of vertebrate metallothioneins
Forty-one amino acid sequences representative of the major classes of vertebrates were aligned (Supplementary Fig. 1) and used to derive new insights on the evolutionary history of vertebrate MTs. The accession numbers of the final set of sequences are listed in Table 1. The maximum likelihood tree is shown in Fig. 4. The ML analysis identified two major clades, one of piscine MTs and another comprising MTs of tetrapods. In turn, the latter is divided in two clades, one of which, made of avian MT1 and mammalian MT4 isoforms, emerges basal to the other groups of tetrapod MTs. The phylogeny of the MT isoforms in mammals clearly demonstrates that among them the MT4 isoform is the most ancestral form, whereas the MT1/MT2 isoforms are the most divergent. Finally, the MT3 clusters forming a sister group of the avian MT2/reptilian MT clade. Noteworthy, the topology of amphibian MT cluster is not perfectly equivalent to the topology inferred when the sequences are analysed alone (Fig. 3).
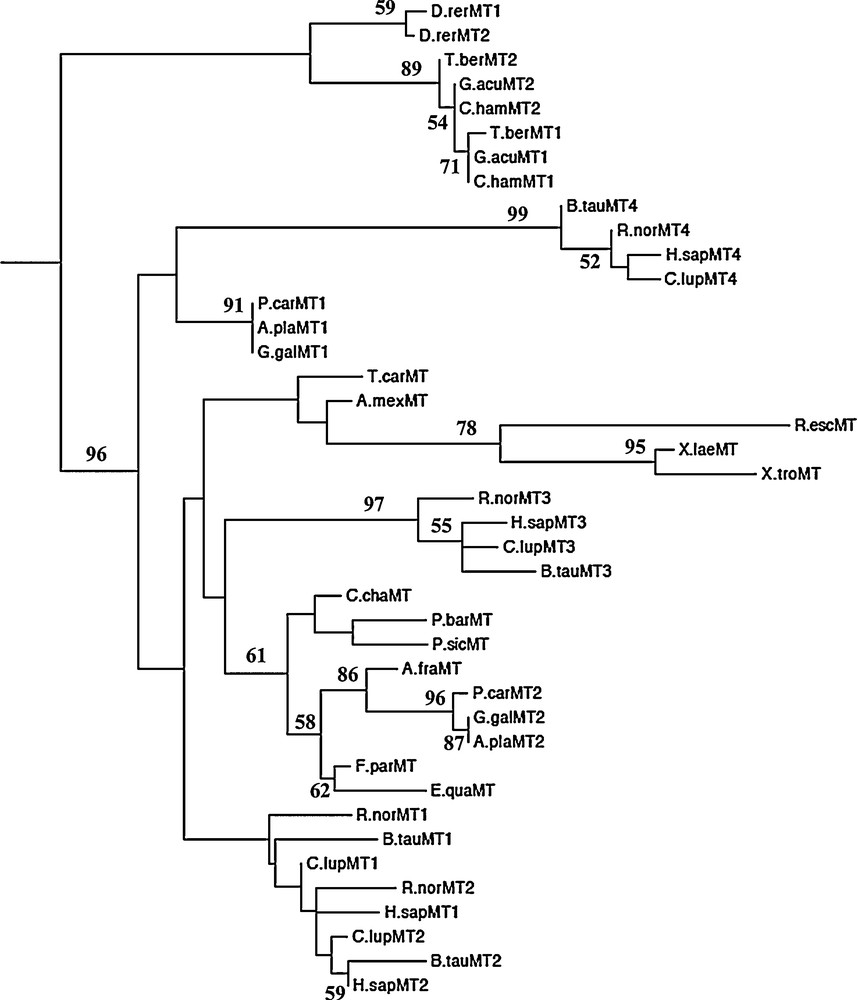
Phylogenetic tree of the amino acid sequences of the vertebrate MT inferred by the Maximum Likelihood (ML) method using the Dayhoff + I + G substitution model implemented in the software SeaView v. 4.0. At the nodes are given values of ML non-parametric bootstrap. For full species name, refer to Table 1.
To better characterize the MTs from the different groups of vertebrates, we calculated the hydropathy indexes of MT proteins used for the phylogenetic analysis; the average values of hydropathy for each group are reported in Table 2. No significant differences in hydropathy values were found between MT1 and MT2 isoforms in fish, birds and mammals; in the latter, however, strong differences were observed among the different MT genes: the average hydropathy was +0.154 for MT1/MT2 isoforms, −0.383 for MT3 and +0.034 for MT4.
Average hydropathy index for the metallothioneins of the different classes of vertebrates.
| Average hydrophaty index | |
| Mammals | |
| MT1–MT2 | +0.154 |
| MT3 | −0.383 |
| MT4 | +0.034 |
| Amphibians | |
| MT | −0.286 |
| Reptiles | |
| MT | −0.169 |
| Birds | |
| MT1–MT2 | −0.187 |
| Osteichthyes | |
| MT1–MT2 | −0.117 |
4 Discussion
Compared with the complexity of MT evolution in invertebrates, the history and fate of MT family members in vertebrates seems to be more straightforward. The huge amount of knowledge gathered regards mammalian MTs, followed by piscine, avian and, more recently, reptilian MTs. The data presented in this paper represent the first comparative analysis on the primary structure of urodelian and anuran MTs. Data also show the phylogenetic relationship between the MT of amphibians and the other vertebrates.
Our results demonstrate an unexpected diversity both in terms of coding and 3′ untranslated regions. MT transcripts from the two species belonging to the genus Xenopus and the axolotl show 3′-UTRs of more than 500 nucleotides. In the axolotl, during embryogenesis three MT transcripts differing only in the length of their 3′-UTRs were described [23]. These transcripts did not arise from three different MT genes, but resulted from events of deadenylation and/or differential polyadenylation signals. In adults, a tissue-specific distribution of two of these transcripts was observed [23]. MT transcripts from the frog R. esculenta and the newt T. carniflex have shorter 3′-UTRs, of about 150 bp. The variability highlighted in the 3′-UTRs could imply differences in stability and translation of amphibian MT transcripts. Indeed, it has been demonstrated that the 3′-UTR offers a diversity of regulatory mechanisms. The untranslated regions control the subcellular localization of transcripts, as well as their translation and stability [30].
Our data record many significant differences also in the primary structure of amphibians MT. A part for the mammalian MT3 and MT4 isoforms, in vertebrates the number of amino acid residues for the MT1 and MT2 isoforms is typical and changeless within the class considered. In amphibians instead the length of the amino acid MT sequence is highly variable, with the three anuran MT displaying 62 amino acids, whereas T. carniflex and A. mexicanum MTs are of 63 and 60 residues, respectively. In particular, A. mexicanum MT lacks three amino acid residues before the first conserved cysteine at N-terminus. Noteworthy, the N-terminal residues upstream of the first cysteine are considered characteristic of the metallothionein origin [31] and the N-terminal region presents the major antigenic epitope [31,32]. Amphibian MTs show also a number of amino acid replacements. The two Xenopodinae X. laevis and X. tropicalis have a histidine residue between the 16th and 17th cysteine. The presence of the histidine residue in the MT sequence is typical of diapsids; it has been proposed that the histidine(s) in the MT sequence plays an important role in metal coordination and stabilization [33].
As for reptilians [19], in the amphibians in so far investigated no MT isoforms have been found. Actually, the paucity of data does not allow us to establish if this condition is typical of the entire class. In addition, the phylogenetic position of the 5 investigated taxa in the gene tree is sill unclear: the phylogenetic tree inferred using as dataset only the amphibian MTs is in contrast with the species tree; however, the presence of the other vertebrate MTs seems to influence the amphibian MT phylogeny. At the moment, we cannot rule out for amphibians a condition similar to that of reptiles, with paralogous genes lost or not sequenced [19,20].
Phylogenetic data presented in this study demonstrate that MT gene duplication in vertebrate species was generally an ancestral event occurred before their speciation; indeed the MT genes are segregated according to isoform and not to species in phylogenetic relationships. The only exceptions are in the piscine clade, where the duplication event seems to follow the speciation in Cypriniformes.
The phylogenetic analyses also indicate that mammalian MT4 is an ancestral form of MT. These results are in agreement with previous observations according to which the MT duplication in mammals occurred before their radiation [10,11]. In their studies about the phylogeny of the MT family in mammals, Moleirinho and coworkers [11] identified the MT4 as the most ancient MT form and hypothesized that it was lost in other tetrapods. Our analysis demonstrates that the avian MT1 is more ancient of the avian MT2 and clusters as a sister group of the mammalian MT4. Avian MT2, together with the paralogous reptilian MTs, forms a unique clade linked to the mammalian MT3 cluster.
The hydropathy index is a parameter inversely proportional to protein flexibility. A higher flexibility can facilitate the conformational changes necessary to the protein for maintaining functionality at low organismal body temperatures. Our previous observations demonstrated that MT hydropathy is a character displaying significant phylogenetic dependence and not associated to the thermal regime or to the environmental adaptation of animal species [15,19,34]. The levels of hydropathy measured for the MTs of the different groups of vertebrate used for this study give more validity to phylogenetic data: the hydropathy index of mammalian MT3 is similar to that of the other tetrapods, as well as the value for MT4 is quite different to that of MT1/MT2 isoforms. The low hydropatic value determined for the MT3 is also in agreement with the high degree of flexibility shown by this protein with respect to the other mammalian MT isoforms [35,36].
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgements
The authors wish to thank Prof. Anna Capaldo for providing the liver samples of the newt Triturus carniflex.


