1 Introduction
The Hyrcanus Group Reid (Genus Anopheles, Subgenus Anopheles) is distributed widely from Europe to East and Southeast Asia, including some of the off-lying islands of the Indian and Pacific Oceans, and at least 27 species are reported within it [1]. In Thailand, eight species members of the Hyrcanus Group have been reported so far, i.e., Anopheles argyropus, Anopheles crawfordi, Anopheles nigerrimus, Anopheles nitidus, Anopheles paraliae, Anopheles peditaeniatus, Anopheles pursati, and Anopheles sinensis [2–4]. Among these, An. nigerrimus, An. peditaeniatus and An. sinensis are considered as suspected vectors of malaria due to Plasmodium vivax [5–9], while An. sinensis and An. peditaeniatus have been incriminated as vectors of P. vivax in China and Korea [10,11] and Japanese encephalitis virus in China and India [12–14], respectively. Even though An. peditaeniatus has been found abundantly and widely distributed throughout Thailand [3,15], its status as a vector of the Japanese encephalitis virus is still a cryptic question, which needs to be investigated more intensively. Recently, An. sinensis and An. nigerrimus have been incriminated as a main vector and secondary or incidental vector, respectively, of Wuchereria bancrofti in Asia [16]. In addition, An. peditaeniatus, An. crawfordi, An. nigerrimus, An. argyropus and An. pursati were reported as high potential vectors of nocturnally subperiodic Brugia malayi [17]. Likewise, the Anopheles hyrcanus group was also considered as an economic pest of cattle because of its vicious biting behavior and ability to transmit cervid filariae of the genus Setaria [2,3].
Establishment of anopheline colonies is the backbone of mosquito researches, and the inability to create a healthy colony of difficult-to-rear species is the principal cause behind every failure in research efforts. Very few research experiments concerning the Hyrcanus Group have been documented, during the past two decades, particularly those with a complete multidisciplinary approach (combination of related-aspects of morphology, cytology, molecular investigation, hybridization, susceptibility and refractory to pathogens, etc.), although eight species members of the Hyrcanus Group are found throughout Thailand and/or other Southeast Asian countries [2,3]. This might result from the lack of biological information and/or available laboratory-raised colonies, particularly the adaptive stenogamous colonies that are easy to maintain and mass produce, which reduces time, workload and manpower for artificially mating adult females with males. Hence, this paper reports establishment of a stenogamous colony of An. peditaeniatus that has existed for more than 20 successive generations, as well as promising possible stenogamous colonies of An. paraliae and An. nigerrimus, which are still being established and detailed. Furthermore, the utility of DNA barcoding, which is incorporated with the taxonomic key for exact identification of the eight species members, is present as well.
2 Materials and methods
2.1 Mosquito species and strains
Eight species members of the Hyrcanus Group were collected in five provinces of western and southern Thailand, where malaria and filariasis are endemic due to Plasmodium falciparum and P. vivax, and W. bancrofti, respectively [16,18]. The species and strains were as follows: An. argyropus (Nakhon Si Thammarat strain: 08°29′N, 100°0′E), An. crawfordi (Trang strain: 07°33′N, 99°38′E), An. nigerrimus (Songkhla strain: 07°13′N, 100°37′E), An. nitidus (Phang Nga strain: 08°27′N, 98°31′E), An. paraliae (Ratchaburi strain: 13°30′N, 99°54′E), An. peditaeniatus (Phang Nga strain: 08°27′N, 98°31′E), An. pursati (Ratchaburi strain: 13°30′N, 99° 54′E) and An. sinensis (Chumphon strain: 10°29′N, 99°11′E). Wild caught, fully engorged females of these species members were collected from cow-baited traps.
2.2 Species identification
Identification of wild caught females followed standard illustrated keys [2–4]. Subsequently, identification using intact morphology of eggs, larvae, pupal skins and adult females were performed intensively in F1-progenies of iso-female lines.
2.3 Molecular investigation
In order to guarantee the exact morphological species identification, thus, individual F1-progeny adult female of each iso-female line was performed for DNA extraction and amplification. Genomic DNA was extracted using DNeasy® Blood and Tissue Kit (QIAGEN, Japan). The LCO1490 (5′-GGT CAA CAA ATC ATA AAG ATA TTG G-3′) and HCO2198 (5′-TAA ACT TCA GGG TGA CCA AAA AAT CA-3′) barcoding primers of Folmer et al. [19] were used to amplify the cytochrome c oxidase subunit I (COI) region of mitochondrial DNA (658 bp, excluding primers). Each PCR reaction was carried out in a 20-μL volume containing 0.5 U of Ex Taq (Takara, Japan), 1X of Ex Taq buffer, 2 mM of MgCl2, 0.2 mM of each dNTP, 0.25 μM of each primer, and 1 μL of the extracted DNA. The amplification profile comprised initial denaturation at 94 °C for 1 min, 30 cycles at 94 °C for 30 s, 50 °C for 30 s, and 72 °C for 1 min, and a final extension at 72 °C for 5 min. The amplified products were electrophoresed in 1.5% agarose gels and stained with ethidium bromide. Lastly, the PCR products were purified using the QIAquick® PCR Purification Kit (QIAGEN, Japan) and their sequences directly determined using the BigDye® V3.1 Terminator Cycle Sequencing Kit and 3130 genetic analyzer (Applied Biosystems of Life Technologies, Japan). The sequence data obtained have been deposited in the DDBJ/EMBL/GenBank nucleotide sequence database under the following accession numbers: An. argyropus (Nakhon Si Thammarat strain: AB781747-AB781751), An. crawfordi (Trang strain: AB781752-AB781756), An. nigerrimus (Songkhla strain: AB781757-AB781761), An. nitidus (Phang Nga strain: AB781762-AB781766), An. paraliae (Ratchaburi strain: AB781767-AB781771), An. peditaeniatus (Phang Nga strain: AB781772-AB781776), An. pursati (Ratchaburi strain: AB781777-AB781781) and An. sinensis (Chumphon strain: AB781782-AB781786). The newly COI sequences were also compared with those available in GenBank using the Basic Local Alignment Search Tool (BLAST) available at http://blast.ncbi.nlm.nih.gov/Blast.cgi. Anopheles gambiae (accession number NC_002084) and Anopheles braziliensis (accession number DQ076238) were used as outgroup taxa [20,21]. Sequences were aligned with BioEdit version 7.0.5.3 [22]. Genetic distance was calculated using the Kimura two-parameter (K2P) model [23]. Using the distances, construction of neighbor-joining trees [24] and bootstrap test with 10,000 replications were performed with the MEGA version 4.0 program [25]. Bayesian analysis was conducted with MrBayes 3.2 [26] by using two replicates of 1 million generations with the nucleotide evolutionary model. The best-fit model GTR + I + G were chosen using the Akaike Information Criterion (AIC) in MrModeltest version 2.3 [27]. Bayesian posterior probabilities were calculated from the consensus tree after excluding the first 25% trees as burn-in.
2.4 Rearing procedures
Mosquito rearing procedures for the Thai Hyrcanus Group (swamp-breeders) followed the detailed techniques described by Choochote and Saeung [28]. All of the experiments were performed in the simple insectarium at 27 ± 2 °C, 70–80% relative humidity, and illumination from a combination of natural daylight from a glass window and fluorescent lighting was provided for approximately 12 h a day.
2.5 Establishment of a stock colony
After exact species identification, based on the intact morphology of eggs, larvae, pupal skins and adult females, and molecular investigations in F1-progenies, the stock colonies of the eight anopheline species were established by pooling five iso-female lines of each anopheline species that have been colonized consecutively for more than 10 generations. These stock colonies were used in the investigation of stenogamous behavior throughout the experiments.
2.6 Screening of stenogamous behavior and establishment of natural mating colonies
Mosquitoes of the ninth generation (F9) were used to determine the natural mating ability in a standard 30 × 30 × 30 cm cage. The reason for using this mosquito generation was based on the fact that any mosquito colony, colonized for more than eight consecutive generations, was of adaptive laboratory mosquito-strains, and easily maintained and mass produced for any experiments. Thus, 200 female and 300 male newly emerged mosquitoes of the ninth generation were introduced into the same cage to co-habit for one week following the former procedures of Sucharit and Choochote [29], and Choochote et al. [30]. This provided a density resting surface (or vertical resting surface per mosquito) of 7.20 [31]. Subsequently, the fasted females were allowed to feed on white rat. Five days after feeding, 20 gravid females were allowed to lay eggs for two days in an oviposited-plastic cup [28], and later, the spermathecae of females were investigated for the presence of sperms. All the eggs obtained from natural copulation in the 30 × 30 × 30 cm cage were processed for hatching, larval and adult rearing, and use for establishing the next stenogamous colony. This process was performed repeatedly from generation to generation until the stenogamous colony was stable.
3 Results
3.1 Species identification
Species identification, based on molecular analyses (cytochrome c oxidase subunit I barcoding region, 658 bp), was generated from a total of 40 specimens of the F1-progenies of the eight species, i.e., using one specimen from each of the iso-female lines of each species. The NJ and Bayesian trees revealed eight distinct clusters. Each species (n = 5) showed a low level of mean intra-specific genetic distances (0.001–0.009), whereas the mean inter-specific genetic distances were high (0.032–0.077) (Table 1). Among the species examined, the NJ tree revealed two major clades. Clade I comprised An. crawfordi, An. paraliae, An. peditaeniatus, and An. sinensis as well as a member of the Korean Hyrcanus Group, Anopheles lesteri. In this clade, the five species were separated from each other, with high bootstrap support (98–100%) (Fig. 1). The phylogenetic relationship revealed that An. paraliae was more closely related to An. peditaeniatus (mean genetic distances = 0.038) than that of An. crawfordi (mean genetic distances = 0.041). In addition, An. paraliae showed closer proximity to An. sinensis (mean genetic distances = 0.032) than that of An. crawfordi (mean genetic distances = 0.047). Remarkably, the low level of mean genetic distances (0.011) between An. paraliae and An. lesteri was observed in this study. Clade II comprised An. argyropus, An. nigerrimus, An. nitidus and An. pursati, which were monophyletic with high bootstrap support (99–100%). Within this clade, An. nitidus was more closely related to An. pursati (mean genetic distances = 0.039) than that of An. nigerrimus (mean genetic distances = 0.049). Additionally, An. nitidus was closely related to An. argyropus (mean genetic distances = 0.054) than that of other species. Anopheles crawfordi and An. pursati were most genetically diverse, with mean genetic distances of 0.077. The Bayesian tree showed almost similar tree topologies to the NJ tree (Fig. 2). Four species members, including An. argyropus, An. nigerrimus, An. nitidus, and An. pursati, were placed within the same clade, with high posterior probability (99%), and well separated from the remaining five species. Anopheles paraliae were nested with An. lesteri with moderate support (posterior probability = 70%), whereas, An. crawfordi, An. peditaeniatus, and An. sinensis was placed into three distinct clusters with high support (posterior probability = 97–100%).
Mean intra- and inter-specific genetic distances (K2P) in eight species members of the Thai Hyrcanus Group based on COI barcoding sequences.
| Species | Mean intra-specific genetic distances (n = 5) | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
| 1. An. argyropus | 0.004 | |||||||
| 2. An. crawfordi | 0.002 | 0.069 | ||||||
| 3. An. nigerrimus | 0.002 | 0.057 | 0.061 | |||||
| 4. An. nitidus | 0.009 | 0.054 | 0.075 | 0.049 | ||||
| 5. An. paraliae | 0.001 | 0.059 | 0.041 | 0.050 | 0.058 | |||
| 6. An. peditaeniatus | 0.005 | 0.069 | 0.057 | 0.067 | 0.059 | 0.038 | ||
| 7. An. pursati | 0.006 | 0.058 | 0.077 | 0.050 | 0.039 | 0.062 | 0.063 | |
| 8. An. sinensis | 0.002 | 0.061 | 0.047 | 0.062 | 0.059 | 0.032 | 0.048 | 0.076 |

Bootstrapped neighbor-joining tree (NJ) of the COI barcoding sequences of eight species members of the Thai Hyrcanus Group and a member of the Korean Hyrcanus Group, Anopheles lesteri. The Anopheles gambiae and Anopheles braziliensis were used as outgroups. Bootstrap values of higher than 70% are shown above the node. Bars represent 0.01 substitutions per site.
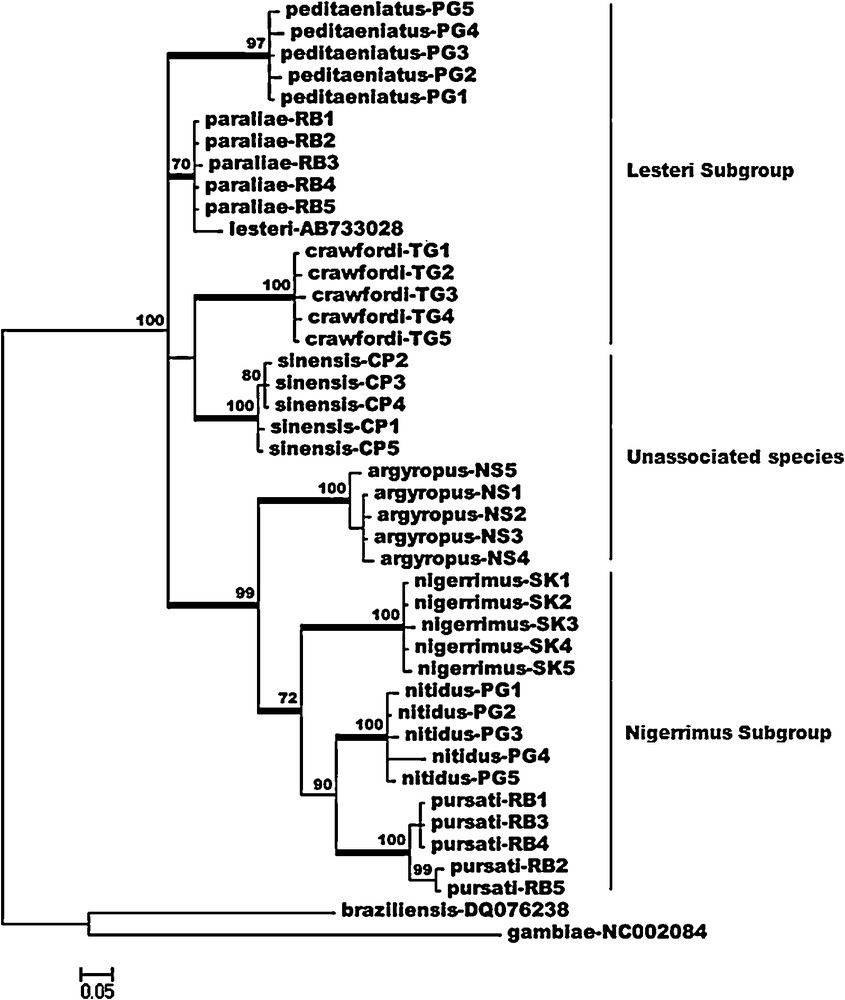
Bayesian phylogenetic tree based on COI barcoding sequences of eight species members of the Thai Hyrcanus Group and a member of the Korean Hyrcanus Group, Anopheles lesteri. The Anopheles gambiae and Anopheles braziliensis were used as outgroups. The posterior probability of higher than 70% is shown above the node. Bars represent 0.05 substitutions per site.
3.2 Establishment of a stock colony
Eight species members of the Hyrcanus Group, i.e., An. argyropus (F23), An. crawfordi (F19), An. nigerrimus (F15), An. nitidus (F16), An. paraliae (F16), An. peditaeniatus (F29), An. pursati (F12) and An. sinensis (F11) were reared successfully for many consecutive generations by using artificial mating techniques. The developmental time of one generation (from eggs to eggs) was more or less 28 days, comprising a duration of 3–4 (3), 8–14 (11) and 1–3 (2) days of egg, larval and pupal development, respectively, as well as added durations of 5-day-old adult females for blood feeding, 5 days for gravid-developed females, and 2 days for egg-laid females.
3.3 Screening of stenogamous behavior and establishment of natural mating colonies
The results of induced natural copulation in a 30 × 30 × 30 cm cage by co-habiting 200 and 300 newly emerged female and male F9 mosquitoes for 1 week, indicated that only five species, i.e., An. pursati, An. sinensis, An. nigerrimus, An. paraliae and An. peditaeniatus could lay eggs successfully with insemination rates of 31%, 33%, 42%, 50% and 77%, respectively (Table 2). The remaining three species of An. argyropus, An. crawfordi and An. nitidus failed to copulate naturally and provided a 0% insemination rate at F9, F10 and F11. After selection, the stenogamous colony of An. peditaeniatus was colonized continuously for more than 20 generations, with insemination rates that ranged from 61–86%. As for An. sinensis, An. pursati, An. nigerrimus and An. paraliae, selection of the stenogamous colony is still in progress for 1, 2, 5 and 6 selected generations of passages, respectively, by yielding the insemination rates ranging from 33–34%, 27–31%, 42–58% and 43–57%, respectively.
The insemination rates of selective stenogamous colonies of Anopheles peditaeniatus, Anopheles paraliae, Anopheles nigerrimus, Anopheles pursati and Anopheles sinensis.
| Mosquito species | Insemination rates of each selected generation of passage (F9–29)a | ||||||||||||||||||||
| F9 | F10 | F11 | F12 | F13 | F14 | F15 | F16 | F17 | F18 | F19 | F20 | F21 | F22 | F23 | F24 | F25 | F26 | F27 | F28 | F29 | |
| An. peditaeniatus | 77 | 81 | 75 | 69 | 62 | 83 | 86 | 76 | 61 | 72 | 68 | 64 | 67 | 85 | 66 | 71 | 80 | 72 | 76 | 74 | 79 |
| An. paraliae | 50 | 49 | 43 | 45 | 57 | 52 | 56 | IP | |||||||||||||
| An. nigerrimus | 42 | 44 | 51 | 46 | 54 | 58 | IP | ||||||||||||||
| An. pursati | 31 | 27 | 30 | IP | |||||||||||||||||
| An. sinensis | 33 | 34 | IP |
a Examined from 100 spermatecae.
4 Discussion
Prior to the study on biology, ecology and behavior of mosquito-borne disease vectors, accurate species identification is the main key for success in obtaining robust information. Consequently, ribosomal (ITS2) and mitochondrial (COI and COII) DNA have been used intensively and widely in the Oriental region for recognition of the species members of the Hyrcanus Group [1,32–42]. Nonetheless, information on COI barcoding sequences for identification of eight species of the Hyrcanus Group in Thailand is lacking. Thus, we initiated this approach to develop DNA barcodes for this species group. Our phylogenetic analyses here separated eight species members of the Thai Hyrcanus Group into eight distinct clusters. In accordance with Harbach [43] and Rattanarithikul et al. [9], An. crawfordi, An. paraliae, and An. peditaeniatus were classified into the Lesteri Subgroup, while An. nigerrimus, An. nitidus and An. pursati were classified into the Nigerrimus Subgroup. The An. argyropus and An. sinensis were categorized into the Unassociated Species. However, interestingly, An. sinensis and An. argyropus show closer relationship to the Lesteri and the Nigerrimus Subgroup, respectively (Figs. 1 and 2). Furthermore, the intra-specific genetic distances was less than 1%, while the inter-specific genetic distances was greater than the 2% value obtained from this study, which is in agreement with the threshold value for distinguishing species based on the COI barcode sequences [44–47]. In contrast to the previously reported of the Neotropical Anopheles (Nyssorhynchus) by Foster et al. [48], our results showed a non-overlapping of intra- and inter-specific genetic distances based on COI sequences. In addition, the COI sequences of An. paraliae, An. peditaeniatus and An. sinensis showed little difference (mean genetic distances = 0.002–0.005) from previous deposited-sequences in GenBank, i.e., An. paraliae (accession number AB733031) [49], An. peditaeniatus (accession numbers AB539069 and AB715091) [42], and An. sinensis (accession number AY444351) [33]. It was interesting to note that An. lesteri from Korea (accession number AB733028) showed a low level (0.011) of inter-specific genetic distances with our An. paraliae, which was congruent with the current results of Taai et al. [49], who suggested that these two morphological species were presumed conspecific, based on results of no post-mating reproductive isolation from hybridization experiments, together with the molecular data of low pairwise genetic distances of COI (0.007–0.017). In conclusion, our findings on mitochondrial DNA sequences of the COI barcoding region (658 bp) could be used for the identification of eight species members of the Thai Hyrcanus Group.
It has long been known that the anopheline mosquito has difficulty in copulating naturally under laboratory conditions, especially in a small space-cage, such as a 30 × 30 × 30 cm cage. However, some species could succeed in copulating in a small space-cage, e.g., Anopheles quadrimaculatus [50,51], An. gambiae complex [52], Anopheles earlei [53], An. sinensis [54–56], Anopheles farauti No. 1 [57], Anopheles albimanus [58], Anopheles subpictus [59], Anopheles balabacensis (Perlis Form = Anopheles dirus B, recently as Anopheles cracens) [29,60], Anopheles annularis [30], An. dirus [61], Anopheles barberi [62], Anopheles sergentii [63], Anopheles freeborni [64], Anopheles barbirostris [65], Anopheles minimus [66], Anopheles albitarsis [67], Anopheles maculatus [68], Anopheles aquasalis [69], Anopheles stephensi [70] and Anopheles pseudopunctipennis [71]. Therefore, artificial mating techniques have been developed by previous investigators in order to solve the mating problems for maintaining laboratory colonies [72,73].
The mating behavior of anophelines in nature usually starts from the males, dancing in a swarm at dusk, often over the tops of bushes and other objects, whereas females may be seen to enter these swarms in small numbers. Each female is grabbed promptly by a male, which locates her through the hearing organs in his antennae, and then, the couple can be seen to fall out from the swarm [2]. In An. maculatus, swarming is 15–21 feet from the ground, which is rather high in comparison to other anopheline species [74]. Additional mating behavior results of Anopheles stephensi var. mysorensis in nature revealed approximately 400 copulations per swarm of 500–600 males, thus, confirming that anopheline females are mated as a result of entering swarms of males [75]. In laboratory conditions, the limited space in a 30 × 30 × 30 cm cage appears to prevent swarming, and therefore causes failure of copulation. However, there are many species of Aedes, Culex and Mansonia mosquitoes that can copulate without males forming a swarm, and they mate easily in small spaces [74,76,77]. In this study, three species members of the Hyrcanus Group (An. argyropus, An. crawfordi and An. nitidus) failed to copulate naturally in a 30 × 30 × 30 cm cage (experiment repeated three times). The success in selecting a stenogamous An. peditaeniatus colony strongly suggested that an artificially mated colony of this anopheline species could be adapted easily to natural copulation in a 30 × 30 × 30 cm cage.
It has been documented that the behavioral polymorphism stenogamy/eurygamy of anophelines is inherited and obviously controlled by one or more genes located on the Y-chromosome [78]. Additionally, the difference of male genitalia morphometry and frequency of clasper movements, which may be involved in the stenogamous behavior of mosquitoes, e.g., between stenogamous An. balabacensis (Perlis Form) and eurygamous An. dirus (Bangkok Colony Strain = An. dirus A, recently as An. dirus s.s.) [60], have been reported [29]. The genitalia of An. balabacensis (Perlis Form) are larger than that of An. dirus (Bangkok Colony Strain) and the former has a shorter duration of clasper movement and mating time than the latter species.
In conclusion, successful selection of the stenogamous colony of An. peditaeniatus, which yielded high insemination rates within a few generations, warrants intensive investigation of a specific stenogamy/eurygamy control mechanism(s) in this anopheline species group, and all the experiments are currently progressing.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgements
This work was supported by the Thailand Research Fund (TRF Senior Research Scholar: RTA5480006) and the Diamond Research Grant of Faculty of Medicine, Chiang Mai University to W. Choochote and A. Saeung. The authors would like to thank Dr Wattana Navacharoen, Dean of the Faculty of Medicine, Chiang Mai University, for his interest in this research.


