1 Introduction
Rice (Oryza sativa L.) confronts several major and minor diseases, among which blast caused by Magnaporthe oryzae [1] is a serious constraint causing moderate to huge economic losses worldwide [2–4]. Outbreaks of this disease are a serious and recurrent problem, and are extremely difficult to control in all rice-growing regions of the world, including Bangladesh [5,6]. Bangladesh experienced several epidemic outbreaks of blast disease since 1980 [7], presumably because of the popularity of certain susceptible cultivars and/or generation of new pathogenic races. Most of the fragrant varieties are highly susceptible to this disease under favourable environment.
There are some chemicals available on the market to control this disease, but they are costly for resource-poor farmers, and sometimes effective chemical control is difficult in case of outbreaks, especially during neck blast. In addition, reduction of chemical application is also desirable for environmental protection in such a heavily farmed country as Bangladesh [8]. Cultivars resistance to blast is therefore the most eco-friendly and economic approach for managing blast diseases in rice. However, cultivars released as resistant often show high levels of susceptibility within a few years, even shortly after cultivar release, due to inadequate information of genotypes used as resistant sources and also due to a lack of information about the pathogen population structure. So, knowledge on population genetics of rice genotypes is important for breeding for blast disease resistance. Though cultivation of resistant varieties is the most promising method to control blast, the breeding-for-resistance programme is still at an early developmental stage in Bangladesh [9].
Fortunately, the Bangladesh Rice Research Institute (BRRI) has already collected more than 8000 rice germplasms. We believe that there are some resistant aromatic or fragrant sources in those materials that may contain blast-resistant gene(s) suitable for developing durable blast-resistant fragrant rice varieties. Recently, in collaboration with the Japan International Research Centre for Agricultural Sciences (JIRCAS), BRRI has developed good research facilities and started an international collaborative study under the “Blast Research Network for Stable Rice Production” project. In connection with this work, the present studies were undertaken with two major objectives: (1) to screen out the aromatic germplasms that are resistant against blast isolate and (2) to confirm the major blast-resistant genes in selected aromatic germplasms using tightly linked molecular markers for the development of durable blast-resistant rice varieties.
2 Materials and methods
2.1 Collection, isolation, purification, and preservation of blast isolate
Rice panicles of high yielding fragrant variety (BRRI dhan34) showing typical blast symptoms were collected from the rain-fed lowland ecosystem of Bangladesh. To obtain a single conidial isolate, an individual conidium was identified with a compound microscope (Olympus BX41) and aseptically transferred into a water agar medium. Finally, the fungus was grown on sterile filter paper and stored at −20 °C after aseptic drying, necessary for repeated access to original isolates [10,11].
2.2 Inoculum preparation and inoculation
To produce inoculum, the stock isolate (paper disc) of single-spore culture was re-cultured in an oatmeal agar medium. The inoculated plates were incubated at temperatures from 25 to 28 °C for 12 to 14 days. The culture was scraped with a sterilized tooth brush and the plates were exposed to continuous light for 4–5 days to induce heavy sporulation. Conidia were dislodged by gentle rubbing with a paintbrush from the incubated plates to sterilized distilled water with 0.01% Tween 20. Spore suspensions were filtered through four layers of gauze mesh and concentration was adjusted to 105 conidia per millilitre using a haemocytometer.
Inoculation of 140 test materials, including 114 fragrant germplasms, 25 differential varieties (DVs) harbouring 23 blast-resistant genes [12–14] and LTH (universal susceptible check) was carried out following the standard methods [10,11]. The seeds of DVs were collected from JIRCAS, Tsukuba, Japan. Inoculated seedlings were incubated in a dew chamber at 25 °C for 20 h, then transferred into a greenhouse maintained at 25 ± 1 °C with 70 to 80% of relative humidity.
2.3 Disease assessment and data collection
Disease reactions of the inoculated plants were evaluated seven days after inoculation using a 0 to 5 scale, where 0 = no evidence of infection, 1 = brown specks smaller than 0.5 mm in diameter, no sporulation, 2 = brown specks about 0.5 to 1 mm in diameter, no sporulation, 3 = roundish to elliptical lesions about 1 to 3 mm in diameter with a grey centre surrounded by brown margins, lesions capable of sporulation, 4 = typical spindle-shaped blast lesions capable of sporulation, 3 mm or longer with necrotic grey centres and water-soaked or reddish brown margins, little or no coalescence of lesions, and 5 = lesions as in 4 but about half of one or two leaf blades killed by coalescence of lesions. The reactions of plant were further categorized as 0 to 2 as resistant (R), whereas 3 to 5 as susceptible (S) in most cases. The scores 2 to 5 were only categorized as susceptible for the differential variety of IRBL5-M and 4 to 5 for IRBLsh-B and IRBLta2-Pi [11,15]. The disease scoring data were then converted into the percent disease index (PDI) by using the following formula [16]:
Cluster analysis was done using Ward‘s hierarchical method using JMP 7.0.2 (JMP Statistics and Graphic Guide, Version 7.0.2: SAS Institute, Inc., Cary, NC, USA) based on a PDI of 140 tested materials. Canonical variant analysis was done using GenStat 5.3 software.
2.4 Confirmation of target genes in selected germplasms by molecular assay
2.4.1 Plant materials
Sixteen selected germplasms, four differential varieties (IRBLsh-B, IRBLta, IRBL9-W and IRBLta-2) that showed low PDI with blast isolate and one susceptible check (LTH) were used for the PCR assays. The plants were grown in a seedling tray for DNA isolation.
2.4.2 DNA extraction and PCR assay
DNA was isolated from leaves collected from 30-day-old seedlings as per the method of Murray and Thomson [17], with modifications. Polymerase chain reaction (PCR) was performed using gene-specific primers (Table 1). The primers of useful genes for Bangladesh conditions were selected from the reaction pattern of tested blast isolate with differential varieties. The PCR mix has a total volume of 10 μL; it contains 25 ng of DNA template, 10 μM of each primer, 25 mM of MgCl2, 10 mM of dNTP, and 0.2 μL of Taq polymerase. The PCR amplification conditions were 1 cycle at 94 °C for 5 min, followed by 35 cycles at 94 °C for 45 s, at 55 °C for 45 s and 72 °C for 90 min, with a final extension at 72 °C for 10 min (G-Storm GS I thermal cycler). The PCR products were detected using 1.5% agarose gel electrophoresis and observation was recorded with a gel documentation system (Alpha Imager EP, Alpha Innotech, CA, USA). The PCR assays were repeated twice for confirmation purpose.
Characteristics of primer used for blast resistant gene identification in selected germplasms.
| Sl. No. | Target gene | Primar name | Size (base) | Primer sequence | References |
| 1 | Pi9 | 195R-1 | 20 | ATGGTCCTTTATCTTTATTG | [18] |
| 195F-1 | 19 | TTGCTCCATCTCCTCTGTT | |||
| 2 | Pita-2 | RM155F | 22 | GAGATGGCCCCCTCCGTGATGG | [19] |
| RM155R | 22 | TGCCCTCAATCGGCCACACCTC | |||
| 3 | Pish | AOL51F | 23 | AGCTGCAGTAGTGCTGTTCCATG | [20] |
| AOL52R | 20 | CGTAAGATCATGAGCGAATG | |||
| 4 | Pita | Pita403F | 20 | CAATGCCGAGTGTGCAAAGG | [21] |
| Pita403R | 20 | TCAGGTTGAAGATGCATAGC |
2.5 Yield and yield contributing characters of selected germplasms
The yield and yield-contributing characters of the selected germplasms were also evaluated at the Field of Plant Pathology Division, BRRI, Gazipur, Bangladesh during the rain-fed season (July–November 2012). Panicle length, filled–unfilled grain per panicle, and 1000 grain weight were collected during harvesting. Principal component analysis was done using PAST multivariate software.
3 Results
3.1 Reaction of blast isolate to differential varieties
The percent disease indexes (PDI) of the tested blast isolate to 25 differential varieties (DVs) harbouring 23 resistant genes Pish, Pib, Pit, Pia, Pii, Pi3, Pi5(t), Pik-s, Pik-m, Pi1, Pik-h, Pik, Pik-p, Pi7(t), Pi9, Piz, Piz-5, Piz-t, Pita-2, Pita, Pi12(t), Pi19, Pi20(t) and LTH are presented in Fig. 1. Among the tested DVs, twelve DVs having Pia, Pii, Pi3, Pi5(t), Pik-s, Pik-m, Pi1, Pik-h, Pik, Pik-p, Pi7(t), Piz-t genes, and LTH showed high PDI (> 75%), eight DVs having Pib, Pit, Piz-5, Pi12(t), Pita, Pi19, Piz and Pi20 genes showed intermediate reactions (50–75%), one DV having Pish gene showed PDI < 50%, and the remaining three DVs having Pi9, Pita-2 and Pita genes showed lower (< 25%) PDI. DVs with low PDI showed four genes, Pish, Pi9, Pita-2 and Pita, which might be the effective ones for developing a blast-resistant fragment rice variety.
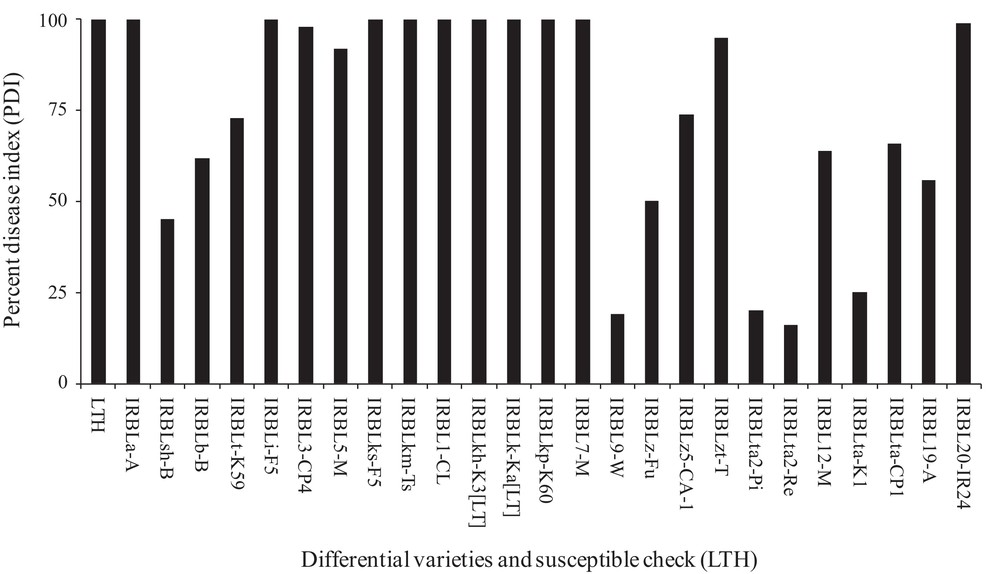
Percent disease index (PDI) of tested blast isolate against differential varieties harboring 23 blast resistant genes and susceptible check (LTH).
3.2 Clustering of genotypes
A dendrogram was constructed using a distance matrix which was calculated from percent disease index values (Supplementary Table S1). This dendrogram resolved the relationship among the 140 tested materials and grouped them into five clusters (Fig. 2). The distribution pattern indicated that cluster I was comprised of the highest number of tested entries (63), followed by cluster V (29), and cluster IV (26). Clusters II and III contained eleven entries each. Among the tested materials, susceptible check ‘LTH’ was placed in cluster V.
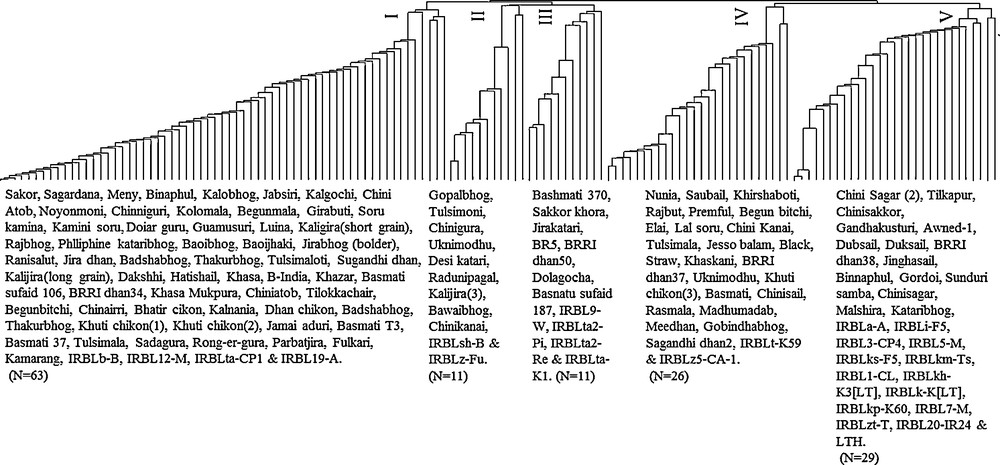
Phenogram of 140 germplasms including fragrant, differential and susceptible varieties. The dendogram was constructed depending on the reaction pattern of germplasms to blast isolate.
3.3 Canonical variant analysis (CVA)
Canonical Variant Analysis was performed to obtain the inter-cluster distances (Mahalanobis's D2 value). The values of inter cluster distance (D2) are presented in Table 2. Statistical distances represented the index of genetic diversity among the clusters. The inter-cluster distance was maximum between cluster III and cluster V (47.28), followed by the distance between cluster II and cluster V (34.57). The minimum inter-cluster distance was observed between cluster II and cluster III (11.17).
Inter cluster distances (D2) of 140 germplasms including aromatic, differential and susceptible varieties.
| Cluster | I | II | III | IV | V |
| I | 0.00 | ||||
| II | 11.53 | 0.00 | |||
| III | 12.71 | 11.17 | 0.00 | ||
| IV | 23.40 | 22.69 | 35.41 | 0.00 | |
| V | 23.88 | 34.57 | 47.28 | 11.87 | 0.00 |
The mean PDI values of five clusters are presented in Fig. 3. The lowest mean value was found in cluster III, followed by cluster II. The genotypes under cluster II and III were resistant to blast disease. The highest mean value was found in cluster V, followed by clusters IV and I.
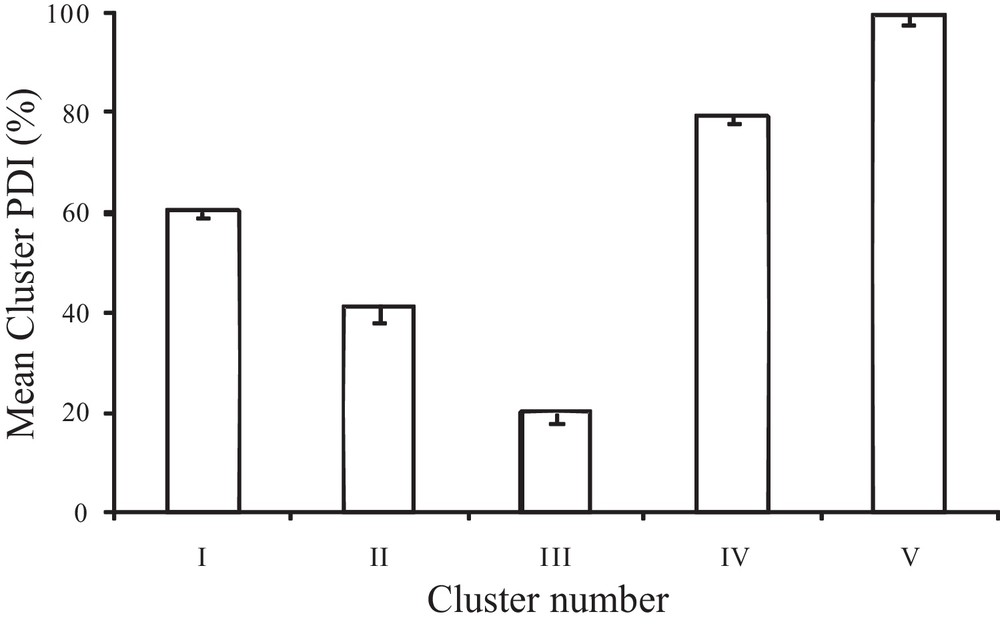
Mean cluster percent disease index (PDI) of five clusters of 140 germplasms including fragrant, differential and susceptible varieties.
3.4 Confirmation of useful genes in selected germplasms
Molecular analyses of the selected 16 germplasms of clusters II and III were done using a gene-based molecular marker along with resistant (target gene containing DVs) and susceptible material LTH. Germplasms of cluster II and III maintained the lowest cluster mean among the five clusters. The useful genes Pish, Pi9, Pita-2 and Pita resistant against the Bangladeshi blast isolate from fragrant rice were determined by the reactions of the tested blast isolate with differential varieties (Fig. 1). The gel picture for the confirmation of Pish, Pi9, Pita-2 and Pita genes are presented in Fig. 4. Among the selected germplasms, we found only three germplasms (BRRI dhan50, Chinigura, and Bawaibhog) that contained more than one target genes in their genetic background. Another eight germplasms contained only one target gene, either Pish or Pita. The rest of the materials did not contain any target gene(s) in their genetic background, though they showed resistant reaction in pathogenicity tests. None of the materials we found that contain Pita-2 gene in their genetic background (Table 3).
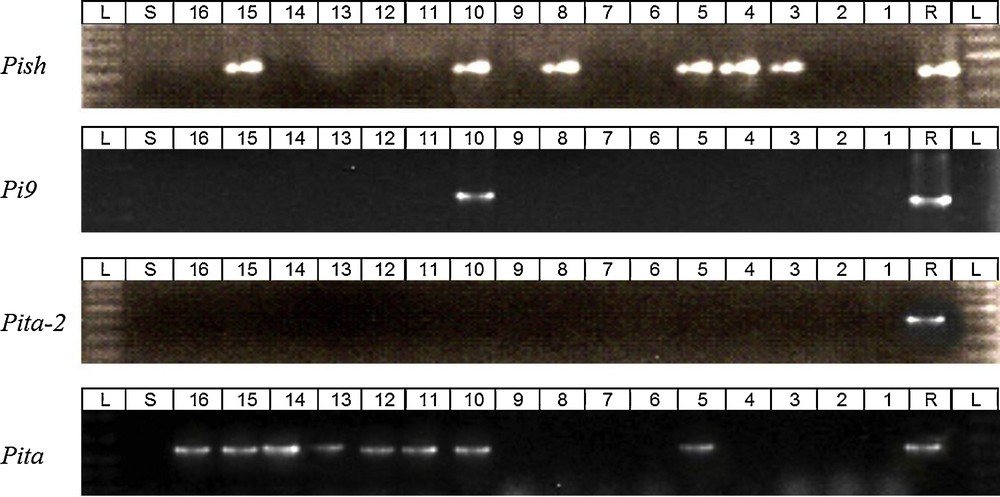
(Colour online). PCR assay of the selected fragrant germplasms for different blast resistance genes (1 = Bashmati 370, 2 = Sakkorkhora, 3 = Jirakatari, 4 = BR5, 5 = BRRI dhan50, 6 = Dolagocha, 7 = Basnatusufaid 187, 8 = Gopalbhog, 9 = Tulsimoni, 10 = Chinigura, 11 = Uknimodhu, 12 = Deshikatari, 13 = Radhunipagol, 14 = Kalijira (3), 15 = Bawaibhog, 16 = Chinikanai, R = differential variety containing respective blast resistance single gene, S = susceptible variety LTH and L = ladder marker).
Frequencies of useful blast resistant genes in selected germplasms.
| Sl. No. | Designation of germplasms | BRRI accession number | Presence of useful genes |
| 1 | Bashmati 370 | 4904 | – |
| 2 | Sakkorkhora | 5347 | – |
| 3 | Jirakatari | 5975 | Pish |
| 4 | BR5 | 4343 | Pish |
| 5 | BRRI dhan50 | 6882 | Pish and Pita |
| 6 | Dolagocha | 451 | – |
| 7 | Basnatu sufaid 187 | 4499 | – |
| 8 | Gopalbhog | 672 | Pish |
| 9 | Tulsimoni | 1980 | – |
| 10 | Chinigura | 4867 | Pish, Pi9 and Pita |
| 11 | Uknimodhu | 5083 | Pita |
| 12 | Deshikatari | 5978 | Pita |
| 13 | Radhunipagol | 6711 | Pita |
| 14 | Kalijira (3) | 247 | Pita |
| 15 | Bawaibhog | 301 | Pish and Pita |
| 16 | Chinikanai | 4356 | Pita |
3.5 Yield potentiality of the selected blast-resistant land races
The results of yield potentiality of the selected resistant germplasms are presented in Table 4. Among these materials, Bashmati 370 provided the highest panicle length (27.7 cm). The sterility percentage of Bashmati 370 was also found in tolerable label (18.4%). The other variety, “Jirakatari”, was also found as long panicle (27.0 cm) and with the lowest sterility percentage (7.3%). The significant variations in grain size were found among these varieties.
Yield contributing characters of blast resistant selected fragrant germplasms.
| Sl. No. | Designation of germplasms | Panicle length (cm) | Filled grain/panicle | Unfilled grain/panicle | Grain sterility (%) | 1000 grain wt. (g) |
| 1 | Bashmati 370 | 27.7 | 110.7 | 20.3 | 18.4 | 18.7 |
| 2 | Sakkorkhora | 22.7 | 85.0 | 61.7 | 72.6 | 10.1 |
| 3 | Jirakatari | 27.0 | 123.0 | 9.0 | 7.3 | 16.0 |
| 4 | BR5 | 22.0 | 221.7 | 105.0 | 47.4 | 16.1 |
| 5 | BRRI dhan50 | 23.3 | 122.3 | 36.7 | 30.0 | 12.3 |
| 6 | Dolagocha | 25.7 | 126.3 | 48.7 | 38.5 | 11.1 |
| 7 | Basnatusufaid 187 | 25.3 | 114.7 | 76.0 | 66.3 | 16.6 |
| 8 | Gopalbhog | 25.5 | 115.0 | 75.5 | 65.7 | 11.6 |
| 9 | Tulsimoni | 26.0 | 170.0 | 72.0 | 42.4 | 7.5 |
| 10 | Chinigura | 23.7 | 153.3 | 17.7 | 11.5 | 23.1 |
| 11 | Uknimodhu | 23.0 | 118.7 | 61.7 | 52.0 | 9.3 |
| 12 | Deshikatari | 23.7 | 107.7 | 58.0 | 53.9 | 8.2 |
| 13 | Radhunipagol | 21.3 | 107.7 | 37.0 | 34.4 | 11.0 |
| 14 | Kalijira (3) | 26.7 | 173.7 | 37.0 | 21.3 | 7.6 |
| 15 | Bawaibhog | 24.3 | 232.3 | 51.3 | 22.1 | 13.9 |
| 16 | Chinikanai | 23.7 | 116.3 | 24.0 | 20.6 | 10.6 |
Three principal components (PCs) accounted for 99.39% of the total variation in the 16 selected fragrant genotypes; of these, the first three PCs exhibited variations of 64.95, 33.82, and 0.61%. The character filled grain had the highest contribution, followed by unfilled grain (Fig. 5).
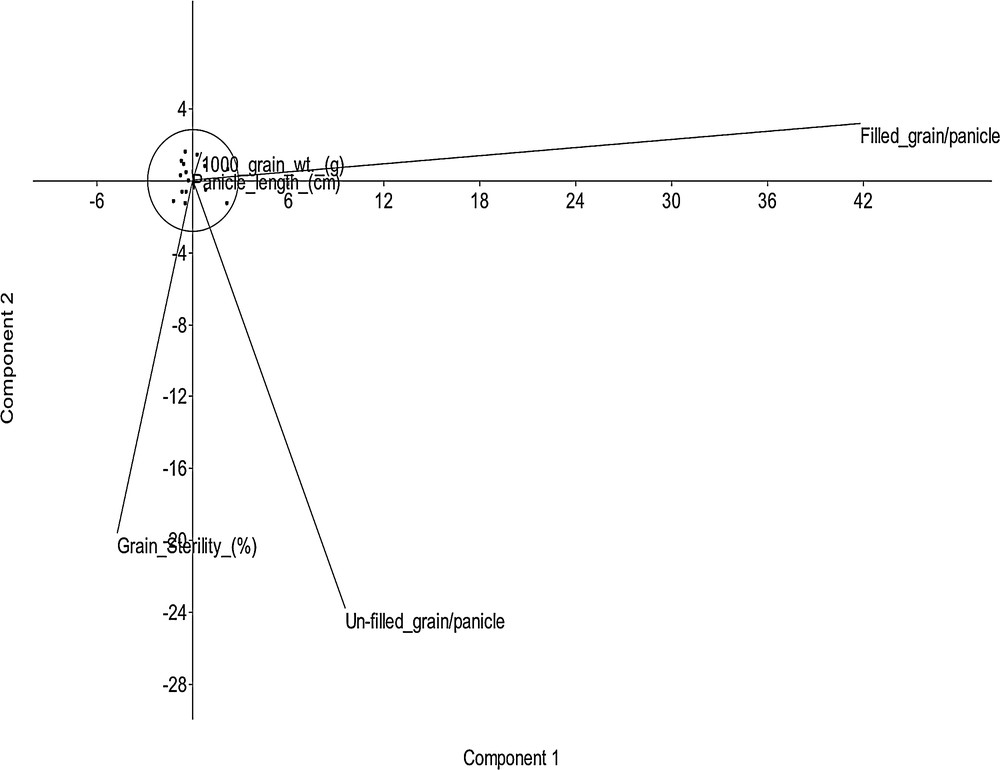
Principal component analysis of 16 selected germplasms based on 5 yield attributing characters.
4 Discussion
Significant variations were found among 140 materials in disease severity to blast, which indicated the existence of a genetic variability of blast resistance in the tested materials. This result was confirmed by the clustering pattern of the germplasms obtained through multivariate analyses. Sixteen fragrant germplasms such as Bashmati 370, Sakkor khora, Jirakatari, BR5, BRRI dhan50, Dolagocha, Basnatu sufaid187, Gopalbhog, Tulsimoni, Chinigura, Uknimodhu, Deshi katari, Radhuni pagal, Kalijira(3), Bawaibhog, Chinikanai, and six differential varieties (DV) such as IRBL9-W, IRBLta2-Pi, IRBLta2-Re, IRBLta-K1, IRBLsh-B and IRBLz-Fu comprised in cluster II and III provided the lowest cluster mean of plant disease index (PDI). It indicated that materials of these clusters may contain blast-resistant useful genes Pish, Pi9, Pita-2 and Pita in their genetic background, because DVs having these similar genes were in the same clusters (Fig. 2). The genotypes having lower PDI values under cluster II and III could be used as parents for the development of blast-resistant varieties. These results are in agreement with those of Latif et al. [22]. The other cluster (V) was comprised of 29 materials, including LTH maintained with the highest cluster mean PDI, which indicated that materials of this cluster are highly susceptible to blast (Fig. 3). The inter-cluster distance between II and III was minimum (D2 = 11.17), indicating that the materials of these clusters were genetically closer. On the other hand, these two clusters maintained maximum distance from cluster V (Table 2).
Genotypes belonging to the distant clusters maintained maximum genetic variation [23]. This variability will greatly help researchers in selecting suitable donors in the breeding-for-resistance programme meant for transferring particular sets of genes into a desirable background [24]. Since the durability of single resistance genes to blast is very limited, efforts in the development of resistant varieties are geared towards stacking of genes to develop gene pyramids. For sustainable utilization of such gene pool for providing an assured resistance against blast, a comprehensive screening of all these accessions against different pathotypes to assess the expression levels of the genes detected is an essential first step for confirmation of useful donors. As expected, the Pita-2 gene was not detected in these accessions; therefore, more rice entries need to be screened to detect this gene. In addition to providing sources of resistance, these germplasms can be extremely useful as the experimental material to develop blast-resistant fragrant rice varieties. Researchers from Japan [25], China [26], India [27], Philippines [28], Thailand [29], and Bhutan [30] screened large number of genotypes and found rice genotypes with resistant genes against blast pathogens. In Bangladesh also, huge numbers of rice entries were screened against blast and resistant entries were identified [31,32]. However, no attempt has so far been made to identify the gene involved, which is essential for maintaining and utilizing the known gene pool.
The results of pathogenicity tests provided important information for selecting useful genes to develop blast-resistant fragrant rice varieties and gene-based markers for molecular study. From the reaction between blast isolate and differential varieties, we found Pish, Pi9, Pita-2 and Pita as being effective against blast isolate. Based on this result, tightly linked gene based marker AOL51 and AOL52 for Pish, RM195 for Pi9, RM155 for Pita-2 and Pita403 for Pita were selected for molecular analyses. Koide et al. [25] carried out a detailed study on the resistance genes and selection DNA markers for blast disease in rice. They found effective markers that were tightly linked to the blast-resistant genes.
5 Conclusion
To identify blast-resistant fragrant sources, the combination of DNA markers and pathogenicity assays is the best way to select donors for the development of durable blast-resistant rice varieties. In this study, only the rice cultivar “Chinigura” contained blast-resistant genes Pish, Pi9 and Pita, while both BRRI dhan50 and Bawaibhog contained Pish and Pita genes in their genetic background. This material may be used as a gene pool to improve blast resistance in rice using marker-assisted or conventional breeding.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgements
The authors are highly grateful to the collaborative research projects between the Bangladesh Rice Research Institute (BRRI) and the Japan International Research Centre for Agricultural Sciences (JIRCAS) entitled “Blast Research Network for Stable Rice Production” (2006–2010) and “Rice Innovation for Environmentally Sustainable Production Systems” (2011–2015) funded by the Ministry of Agriculture, Forestry and Fisheries of Japan for providing all necessary supports.


