1 Introduction
Studies involving abiotic stress tolerance in plants have identified that the expression of many genes vary in response to various abiotic stresses viz. temperature stress (both low and high), drought, salinity, oxidative stress, etc. [1], which resulted in reduced crop productivity [2,3]. Studies carried out for genome wide mRNA abundance have validated that the expression of around 5–30% of the genes are controlled by various abiotic stresses [4,5]. Plants have attained different strategies (mainly adaptive) at morphological, physiochemical, biochemical and cellular levels, which help them survive environmental extremes. Upregulated genes have always been given more importance, and extensive research on members of DREBs, MYB, bZIP proteins, and zinc finger families represents their importance in response to various environmental stresses. However, only a handful of literature [6] is available on genes, which are downregulated and research on all these genes are still lacking behind for elucidating their possible roles in the regulation of plants responses to stress.
Reports on genes encoding chlorophyll a/b-binding proteins, plastocyanin, and PSI subunit proteins have revealed their downregulation in response to cold stress [7–9]. It has been established that the plants acquire increased tolerance to photo-inhibition [10,11] particularly, owing to the shifting of photosynthetic carbon metabolism [12]. Plastocyanin, a type-I copper-containing a protein is found in cyanobacteria, algae, and plant chloroplasts. It is considered as one of the most abundant proteins in thylakoids [13,14] and plays a role in both linear and cyclic photosynthetic electron transport [15]. Plastocyanin, synthesized in the cytosol, acquires its mature form and Cu2+ co-factor [16] in the thylakoid lumen [17]. Light-stimulated responses are causative agents for the increase of plastocyanin mRNA levels and have been reported to play an important role in Arabidopsis, Hordeum, and Nicotiana [18,19]. Arabidopsis, Nicotiana, Oryza and other higher plants have been observed to possess/express two isoforms of plastocyanin [20–23]. PETE2 (DRT112) protein has been preferentially observed to be more abundant than PETE1 and their independent regulation for performing different roles in copper homeostasis as well as electron transport is known [24,25]. Studies on double mutant generated for Arabidopsis plastocyanin genes revealed their failure for growth due to an inhibition in light-driven electron transport [26]. Overexpression of the Spinacia plastocyanin promoter in Nicotiana has revealed a differential regulation of plastocyanin expression by oxidized and repressed plasto-quinone pool [27].
Lepdium latifolium L. ecotype Ladakh shows remarkable ability to withstand harsh environmental stress conditions [28] at different developmental stages, making it an excellent candidate for the exploration of genes related to abiotic stresses. Hence, Lepidium can serve as a source of genes and regulatory elements for abiotic stress tolerance through translational genomics approach and may also further aid in providing a deeper insight into the interaction of genes and networks, clarifying the interaction of genes and regulatory networks under adverse conditions [29]. Here, we describe the isolation, full-length cloning and expression pattern of DNA damage repair/toleration protein from Lepidium latifolium L. in response to various abiotic stresses.
2 Materials and methods
2.1 Plant material
Seeds of L. latifolium L. (Ladakh ecotype) were procured from Leh, India (11,500 ft asl). The collected seeds were germinated and seedlings were grown at 25 ± 2 °C, 150 μmol m−2 s−1 light intensity, 75% relative humidity and a 16:8 h photoperiod [30].
2.2 Rapid amplification of cDNA ends (RACE) and intron mapping
Total RNA was isolated from L. latifolium plants (three-week seedlings) by RNeasy mini kit (Qiagen, Germany) using the manufacturer's protocol from unstressed (control) and stressed leaves (4 °C for 24 h) of L. latifolium, quantified by spectrophotometry (Biorad, USA) and stored at –80 °C until further use. Based on EST sequence (FG618360), gene specific primers using Primer3 [31] were designed for RACE, which was carried out using GeneRacer™ kit (Invitrogen, USA) following the manufacturer's protocol. The amplified fragments were cloned in pGEM-T Easy vector (Promega, USA) and sequenced by M13 universal primers. The sequenced 5′ and 3′ RACE fragments were aligned together for removal of overlapping sequences and generation of full-length LlaDRT from Lepidium. Subsequently, extreme forward and reverse primers were designed from the ends of the full-length DRT gene sequence. All the primers designed for the study of LlaDRT are listed in Table 1.
Description of the oligonucleotides used in this study.
| S. No | Name | Sequence (5′–3′) | Length (mer) |
| 1 | Primers for 3′ RACE | ||
| LlaDRT F1 | CGAGGTTGCCTTGACTGAGC | 20 | |
| LlaDRT F2 | GCCACATCAGGGTGCTGGTA | 19 | |
| GeneRacer™ 3′ outer | GCTGTCAACGATACGCTACGT AACG | 25 | |
| GeneRacer™ 3′ nested | CGCTACGTAACGGCATGACAG TG | 23 | |
| 2 | Primers for 5′ RACE | ||
| LlaDRT R1 | GCCATACAACAATCACGGTCCA | 22 | |
| LlaDRT R2 | CCTCTAATTGAGGCCGAGAC | 20 | |
| LlaDRT R3 | TACCAGCACCCTGATGTGGC | 20 | |
| GeneRacer™ 5′ outer | CGACTGGAGCACGAG GACACT GA | 23 | |
| GeneRacer™ 5′ nested | GGACACTGACATGGA CTGAAG GAGTA | 26 | |
| GeneRacer™ RNA oligo sequence | CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA | 44 | |
| Oligo dT primer | CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA | 60 | |
| 3 | Primers for full-length cloning and intron mapping | ||
| LlaDRT F | ATGGCCTCAGTAACCTCAGCC | 21 | |
| LlaDRT R | TTAGTTAACGGTGAGTTTACCG | 22 | |
| 4 | Primers for qPCR | ||
| 26SrRNA F | CACAATGATAGGAAGAGCCGAC | 22 | |
| 26SrRNA R | CAAGGGAACGGGCTTGGCAGAAT | 23 | |
| LlaDRT F1 | CGAGGTTGCCTTGACTGAGC | 20 | |
| LlaDRT R1 | GCCATACAACAATCACGGTCCA | 22 |
2.3 Multiple alignments and bioinformatics analysis
The full-length LlaDRT (JN214346) cDNA and the deduced amino acid sequence were compared against a non-redundant database of NCBI. Multiple alignments of the retrieved full-length amino acid sequences from the database were generated using ClustalW [32] and a bootstrapped phylogenetic tree [33] was obtained following the neighbour-joining method [34] using MEGA5 software [35]. The conserved motifs in sequences encoded by DRT cDNAs were detected by analysing protein sequences by MEME and BLOCKS software [36]. Conserved domains in DRT encoding sequences were analysed using the online utilities PROSITE (http://web.expasy.org/docs/swiss-prot_guideline.html) and SMART (http://smart.embl-heidelberg.de/).
2.4 Comparative quantification of transcript accumulation by qPCR
To investigate the expression patterns of LlaDRT in different concentrations of abiotic stress treatments (salicylic acid, absciscic acid, jasmonic acid, calcium chloride, and ethylene) at a given point of time, total RNA was isolated from Lepidium plants by RNeasy mini kit (Qiagen, Germany), following the manufacturer's instructions. The treatments of Lepidium seedlings (three weeks) were carried out following the procedure in [37]. cDNA was synthesized from purified (RNase free DNase treatment for genomic DNA contamination removal) RNA and quantified using a spectrophotometer. Equal quantities of synthesized cDNA were subjected to quantitative PCR (Mx3005P Stratagene/Agilent, USA) with SYBR green mastermix (Qiagen, Germany) and gene specific primers for LlaDRT in the reaction set-up. A housekeeping gene (26S rRNA) was used as the reference gene for the normalization/calibration reaction, which was amplified separately. The PCR conditions followed for the reaction set-up were initial denaturation at 95 °C for 10 min followed by 40 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 30 s. The qPCR reactions were performed thrice to assess the reproducibility. Ct values obtained were analysed using statistical program Cropstat version 7.2 (IRRI, Philippines).
3 Results
3.1 Rapid amplification of cDNA ends (RACE)
The first round of 5′ RACE was carried out using the designed primers, which yielded an amplified product of 683 bp using Generacer™ 5′ outer primer and LlaDRT R1. Subsequent nested PCR with diluted template (1:50) of the above reaction with Generacer™ 5′ nested primer and LlaDRTR2 followed by Generacer™ 5′ nested primer and LlaDRTR3 resulted into the amplification of 597 bp and 526 bp, respectively (Fig. 1). In a similar fashion, 3′ RACE was carried out, first with Generacer™ 3′ outer primer and LlaDRT F1, followed by nested amplification using the primer set Generacer™ 3′ nested primer and LlaDRT F2 yielded amplified products of 296 bp and 251 bp, respectively (Fig. 2). The resulting clones obtained from RACE were cloned, sequenced and aligned to obtain full-length LlaDRT (JN214346) and reconfirmed by PCR amplification using LlaDRT F and LlaDRT R primers designed after the ORF prediction.
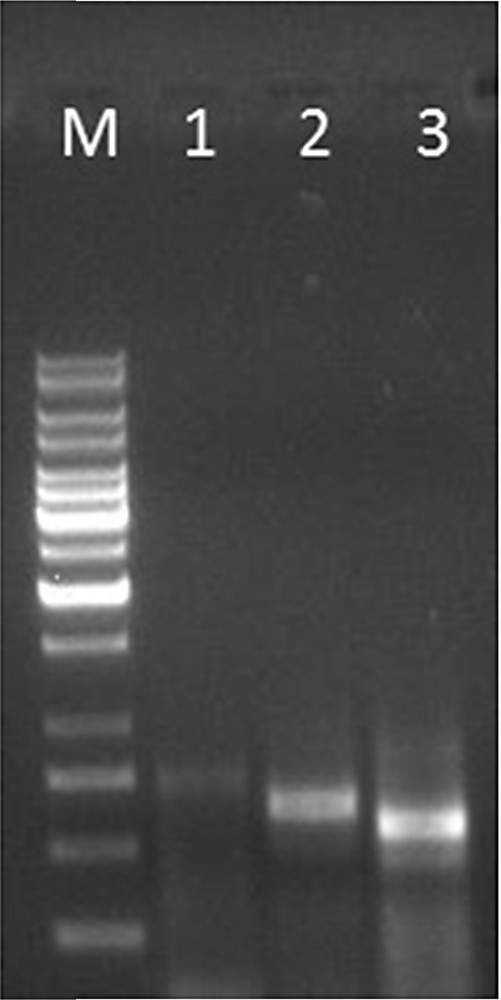
The amplicons obtained after 5′ RACE in Lepidium latifolium L. (M – 1 Kb DNA ladder (Fermentas, Lithuania); L1 – product amplified (683 bp) using LlaDRT R1 and GeneRacer™ 5′ outer; L2 – product amplified (597 bp) using LlaDRT R2 and GeneRacer™ 3′ nested primer; L3 – product amplified (526 bp) using LlaDRT R2 and GeneRacer™ 3′ nested primer).
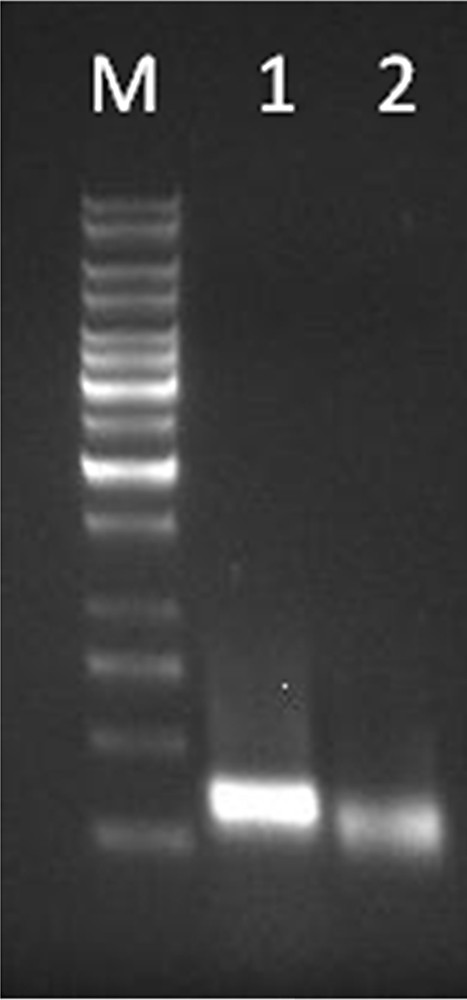
The amplicons obtained after 3′ RACE in Lepidium latifolium L. (M – 1 Kb DNA ladder (Fermentas, Lithuania); L1 – product amplified (296 bp) using LlaDRT F1 GeneRacer™ 3′ outer; L2 – product amplified (251 bp) using LlaDRT F2 and GeneRacer™ 3′ nested primer).
3.2 Intron mapping of LlaDRT in L. latifolium L. Genome
cDNA and genomic DNA were amplified using LlaDRTF and LlaDRTR primers to find out the presence of introns, which revealed a similar product size of 504 bp (Fig. 3), which was confirmed by sequencing. This proves that LlaDRT from L. latifolium L. genome is devoid of introns.
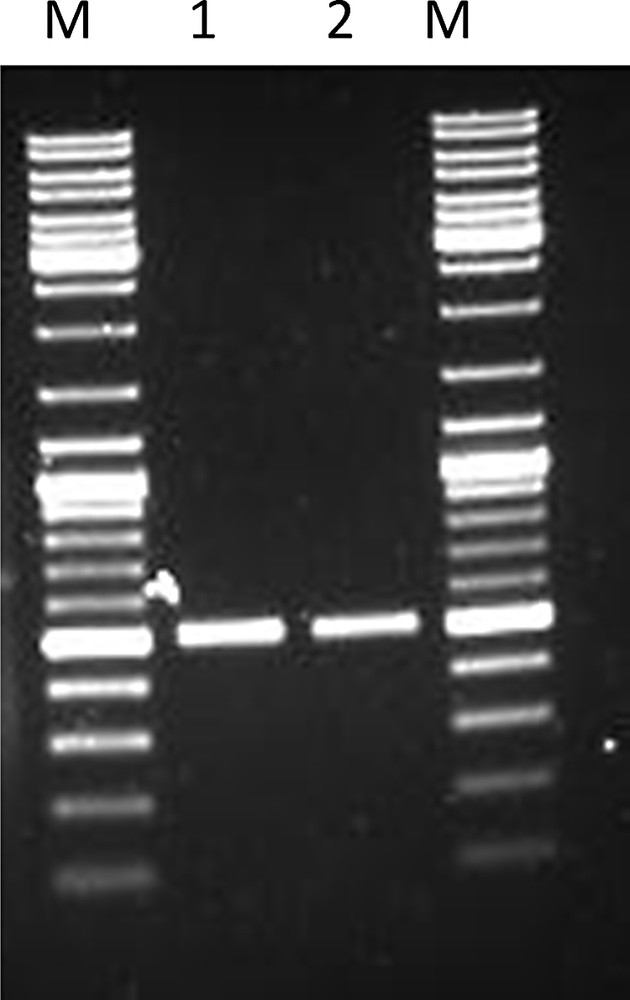
Full-length amplification of LlaDRT from Lepidium latifolium L. (M –1 Kb DNA ladder mix (Fermentas, Lithuania); L1 – product amplified (504 bp) using LlaDRT F LlaDRT R using genomic DNA as a template; L2 – product amplified (504 bp) using LlaDRT F and LlaDRT R using cDNA as a template).
3.3 Bioinformatics predictions for isolated LlaDRT
Retrieval of amino acids sequence using BLAST analyses and subsequent alignment by ClustalW revealed significant homology of LlaDRT with Arabidopsis (86%) and over 60% with most of the other similar protein sequences from dicots and monocots (Table 2). The bootstrapped phylogenetic tree constructed confirmed the closeness of LlaDRT with Arabidopsis, which was likely, since both are members of the Brassicacae family (Fig. 4). The multilevel consensus sequences in related sequences of LlaDRT were detected by MEME and BLOCKS (Table 3, Fig. 5), which indicates a 130-aa long conserved motif stretch with a match of P-value < 0.0001 and e-value < 10, suggesting a high conservation of the plastocyanin gene amongst plant species. Domain analysis elucidated a conserved copper-containing domain (TIGR02656) from 70–149 (cd length of 99) with an e-value of 5.48e–45. The members of this family contain blue copper protein associated/related closely with pseudoazurin, halocyanin, amicyanin, etc., located in the thylakoid lumen [26] and are considered as performing electron transport to photosystem I in cyanobacteria and chloroplasts. Another multidomain PETE (COG3794) was also detected from 73–167 (cd length of 128), having an e-value of 4.56e-16, which signifies the putative role that LlaDRT may play regarding energy production and conversion. The domain and other signatures sequences are probably retained by L. latifolium L. during the course of evolution, surpassing other selective constraints and thus, enabling plants to retain the properties associated with it. However, during the evolutionary processes, divergence from other similar sequences has also been observed by phylogenetic analysis, and it is possible that despite several similarities and conserved nature, it may have additional roles to play.
Similarities in protein sequences of LlaDRT with reported sequences from other plants.
| S. No | Plant | Gene name | Accession no | Length (aa) | % Similarity with LlaDRT |
| 1 | Arabidopsis lyrata | DNA damage repair/toleration protein 112 | XP_002893099 | 167 | 86 |
| 2 | Arabidopsis thaliana | Plastocyanin major isoform | AEE29963 | 167 | 86 |
| 3 | Arabidopsis thaliana | DRT112 | AAA32787 | 167 | 83 |
| 4 | Ricinus communis | Plastocyanin A, chloroplast precursor | XP_002510603 | 168 | 68 |
| 5 | Pisum sativum | Plastocyanin | P16002 | 168 | 67 |
| 6 | Silene latifolia | Plastocyanin, chloroplastic | P07030 | 165 | 67 |
| 7 | Nicotiana benthamiana | Chloroplast plastocyanin precursor | ACV32157 | 169 | 69 |
| 8 | Cucumis sativus | Plastocyanin, chloroplastic like | XP_004136366 | 166 | 70 |
| 9 | Lotus japonicus | Unknown | AFK36672 | 167 | 67 |
| 10 | Populus trichocarpa | Predicted protein, plastocyanin | XP_002307754 | 168 | 62 |
| 11 | Medicago trunculata | Plastocyanin | XP_003603843 | 167 | 66 |
| 12 | Solanum tuberosum | Plastocyanin precursor | AFW90573 | 170 | 66 |
| 13 | Solanum commersonii | Plastocyanin precursor | CAJ19272 | 171 | 62 |
| 14 | Plantago major | Plastocyanin | CAJ34813 | 160 | 61 |
| 15 | Zea mays | Plastocyanin | NP_001147504 | 155 | 63 |
| 16 | Hordeum vulgare | Plastocyanin precursor | CAA68696 | 155 | 53 |
| 17 | Oryza sativa | Plastocyanin precurssor | AAB63590 | 154 | 57 |
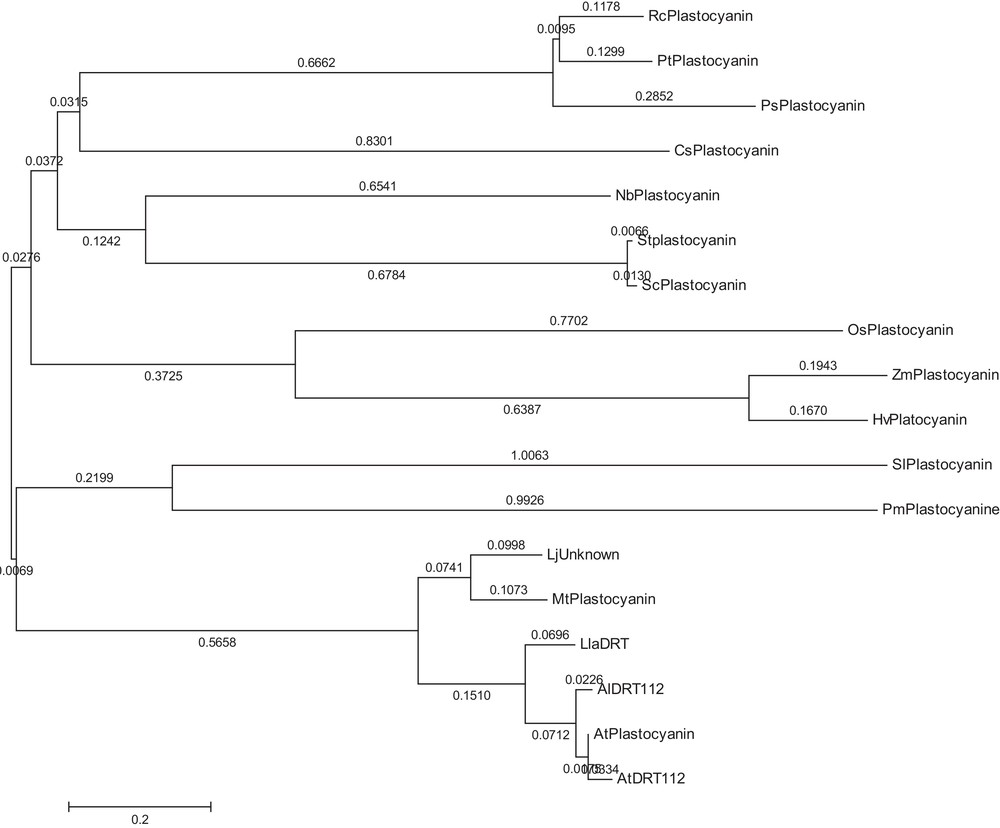
Phylogenetic relationship of LlaDRT in 15 different taxa deduced on the basis of protein sequence similarities.
Multilevel consensus sequences for the MEME defined motifs among different DRT gene sequences. The motif match has a P-value < 0.0001 and the sequences used have an e-value < 10.
| Motif | Width | Multilevel consensus sequence |
| 1 | 130 | PRLCIKASLKDFGVAVVATAASIMLASNAMAIEVLLGGDDGGLAFIPNDFSIAKGEKITFKNNAGFPHNVVFDEDEIPSGVDVSKISMDEQDLLNAPGETYEVTLTEKGTYSFYCAPHQGAGMVGKVTVN |
| 2 | 21 | MATVTSAAVAIPSFTGLKAGR |
| 3 | 64 | LLAGGAMAQDVLLGANDGVLVFEPNDFTVKAGETITFKNNAGFPHNVVFDEDECPSGVDWSKIS |
| 4 | 29 | QEEYLNAPGETYSVTLTVPGTYGFYCEPH |
| 5 | 14 | IKSSATVRIQTAAV |
| 6 | 11 | QGAGMVGKVTV |
| 7 | 11 | RRRRWVVRASL |
| 8 | 14 | KVSAIAKIPTSNSQ |
| 9 | 8 | CSSSRVST |
| 10 | 6 | PKWWIR |

(Colour online.) Motif identified using MEME and BLOCKS.
3.4 Quantitative expression analysis of LlaDRT in response to various abiotic stresses
Temporal gene expression studies of LlaDRT in response to various abiotic stresses were carried out by qPCR as described in the Materials and methods section. Documented studies on the induction of salicylic acid by abiotic stress in mutants and transgenic plants are available [38], and the expression level of the transcript when analysed by qPCR revealed the downregulation of LlaDRT by 0.67-fold and 0.73-fold when exposed to 10 μM and 50 μM salicylic acid stress, respectively compared to the control treatment (Fig. 6a), which may contribute to severe damage in abiotic stress. Considering that salicylic acid does not operate alone but moves in tandem with other phytohormones, like jasmonic acid and ethylene [39,40], we analysed the expression of LlaDRT under jasmonic acid and ethylene treatments. Jasmonic acid-related genes are reported to regulate defence mechanisms during pathogenesis and wounding [41]. Reports are available for jasmonic acid application for reduction of injury caused by cold stress in fruits [42]. Subsiding the salicylic acid operation with jasmonic acid, we also thought that plants might be responding to low temperature stress in a similar way to that they respond to pathogenic response/wounding stresses. qRT–PCR analysis of Lepidium seedlings submitted to 10 μM and 50 μM jasmonic acid foliar sprays showed significant transcript decreases (0.43-fold and 0.20-fold, respectively) compared to that of the control treatment set (Fig. 6b), reflecting its possible role in the freeze tolerance of plants, which may be further examined either by reverse genetics or metabolomics. Transcript accumulation was observed as 0.88-fold and 1.03-fold change by 10 μM and 50 μM ethylene foliar spray (Fig. 6c), respectively, suggesting that the ethylene responsive genes are not affecting LlaDRT and are independent of ethylene responsive pathways for its regulation. ABA is involved in the regulation of diverse plant processes and mediates expression of various stress responsive genes [43,44]. However, studies on Arabidopsis have shown that ABA responsive genes are induced by drought, but seem to be independent of ABA for the cold effect [45]. Seedlings exposed to ABA foliar sprays of 10 μM and 50 μM, when analysed with qPCR, showed significant downregulation of the LlaDRT transcript falling to 0.08-fold and 0.11-fold respectively, in comparison to the Mock/control treatment (Fig. 6d) qPCR carried out on Lepidium seedlings sprayed with 1 mM and 5 mM of calcium chloride, which showed a significant increase of the transcript with a 5 mM foliar spray. The fold change recorded for 1 mM and 5 mM sprays were 1.17 and 5.57 in comparison with the control (Fig. 6e), suggesting that the secondary messenger plays a role in LlaDRT regulation.
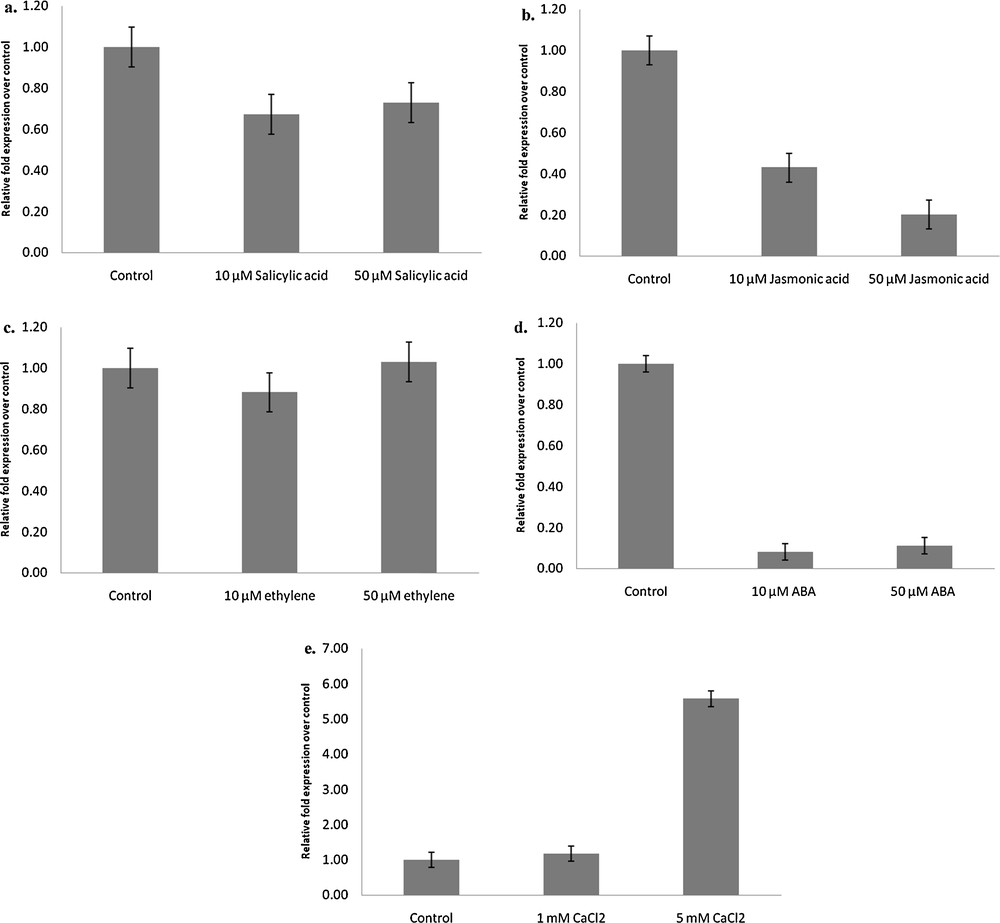
Relative quantitative expression of LlaDRT in response to different abiotic stress regulators, viz. salicylic acid, jasmonic acid, ethylene, absciscic acid and calcium chloride. Lepidium latifolium seedlings exposed to (a) 10 μM and 50 μM of salicylic acid, (b) 10 μM and 50 μM of jasmonic acid, (c) 10 μM and 50 μM of ethylene, (d) 10 μM and 50 μM of absciscic acid, (e) 1 mM and 5 mM of calcium chloride. The treatment of stress regulators was given for 8 h and 26srRNA was used as an internal control. The error bars in each figure represent SE.
4 Discussion
DNA damage repair/toleration protein (DRT112), a copper ion binding/electron carrier, is one of the two reported Arabidopsis plastocyanin genes (PETE1 and PETE2), and is thought to be a predominant form, expressed around ten times stronger than PETE1. PETE2 (DRT) is expected to be post-transcriptionally regulated via copper homeostasis and thought to participate in the electron transfer between P700 and the cytochrome b6f complex in PSI. Physiological, biochemical and molecular changes [10,46] triggered by abiotic responses result from stable and long-term adaptations, explaining complex but necessary developmental responses [46]. Under the influence of low temperatures, inhibition of various metabolic processes, including photosynthetic carbon dioxide assimilation, occurs, which in turn creates imbalance and photo-oxidative damage [47]. Thus, energy imbalance is created; as a consequence of that, plants are forced to downregulate the photosynthetic genes for reduction in the absorbed light energy [48] and its recovery occurs by re-programming of the associated carbon metabolism [12,46]. This study highlights stress specific changes in the transcript level by application of different stress regulators. The increase in level of transcripts may be correlated with carbohydrate-mediated repression of photosynthesis when plants are shifted to a colder environment [48,49]. The fact that the same kind of results was not observed for all the studied abiotic stress treatments reflects that these stresses have different impacts on photosynthesis, both in shorter and longer durations. This indicates that if the studied gene is used for the generation of transgenic or breeding programs, it may provide some interesting regulations that had not yet been unfolded so far. This study also makes a major point in potential values of studying plastocyanin-related genes; the results obtained in the frame of our study may provide useful points for investigating the molecular mechanism of photosynthetic genes regulation during abiotic stress.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgement
Vimlendu Bhushan Sinha is thankful to the Defence Research Development Organization (DRDO), India, for financial support. The authors also thank the Defence Institute of High altitude Research (DIHAR), Leh, India for their cooperation in collection of the plant material.


