1 Introduction
The genus Lolium (ryegrasses) is a member of the Poaceae family, is native to Europe, the Near East and North Africa and consists of nine diploid species altogether with the somatic chromosome number of 2n = 2x = 14 [1,2]. Of which, Italian ryegrass (L. multiflorum) and perennial ryegrass (L. perenne) are cultivated widely in the temperate climate areas of the world as forage and turf grasses [1]. L. rigidum is a winter annual grass originating from the Mediterranean region, and it is not only used as a cultivated fodder crop in some areas, mostly in Australia, adapting to drought and grazing pressure [3], but also an economically damaging crop weed [4] having evolved resistance to many different chemical herbicides [5,6]. The three above-mentioned species are generally self-incompatible species, and therefore each germplasm could be regarded as a heterogeneous population of genotypes [7]. In addition, the previous studies have reported that the Lolium and broad-leaved fescue (Section Schedonorus) are closely related, in particular to tall fescue (Festuca arundinacea), therefore, these grasses are commonly referred to as the Lolium/Festuca complex [8].
Simple sequence repeats markers (SSRs) have been proved to be an ideal tool for genotyping elite grass material, surveying genetic resources, map construction and genetic variability identification because they are codominant, multi-allelic, highly reproducible, abundant and are evenly distributed in the plant genome either in coding and noncoding regions [9,10]. While most of the strategies currently used to isolate SSR markers are both time- and cost-consuming, which limits the application of SSR in those genomes with less information [11]. Therefore, cross-species and cross-genus amplification of molecular markers is now a common strategy for the discovery of markers to use on the not so well studied species [12,13]. It has been reported that SSR markers has showed a high transferability across species or even in closely related genera [14], which originated from genomic libraries (genomic Simple Sequence Repeat, gSSRs) and/or derived from expressed sequence tags (ESTs) on the most important crops or model species, and thus are now frequently used on diversity, evolutionary and mapping studies in other related species [15–18].
At present, a large number of SSR markers have been developed for the Lolium/Festuca species, such as L. perenne [19], L. multiflorum [20] and tall fescue (F. arundinacea), a species closely related to Lolium [21–23]. However, SSR markers in L. rigidum has not been developed. Therefore, the main objectives of our present study are to: (1) assess the transferability of published Lolium/Festuca SSRs cross three Lolium species, including L. rigidum, L. perenne and L. multiflorum, (2) evaluate polymorphism and efficiency of the transferable SSR markers for the genetic diversity and relationship analyses among three ryegrass species, and (3) facilitate genetic studies and molecular breeding for ryegrasses programs.
2 Material and methods
2.1 Plant material
All plant materials including eight accessions from three important species of Lolium were obtained from the National Plant Germplasm System, USA (http://www.ars-grin.gov/) (Table 1). Seeds were germinated and the ploidy was identified as a diploid (2n = 2x = 14).
Accessions of Lolium species used in the analysis.
| Species | Code | Original ID | Origin | Breeding State | Sample size |
| Lolium multiflorum | LM1 | PI 187220 | Belgium | Uncertain | 8 |
| LM2 | PI 239486 | Spain | Uncertain | 9 | |
| LM3 | PI 241913 | Italy | Wild material | 8 | |
| Lolium perenne | LP1 | PI 418725 | France | Wild material | 7 |
| LP2 | PI 619018 | Wales | Wild material | 7 | |
| Lolium rigidum | LR1 | PI 314447 | Georgia | Wild material | 8 |
| LR2 | PI 422586 | Morocco | Wild material | 9 | |
| LR3 | PI 545666 | Turkey | Wild material | 8 |
2.2 Genomic DNA extraction
Total genomic DNA was extracted from leaf tissues, with an average of eight genotypes per accession, by the cetyltrimethylammonium bromide (CTAB) protocol [24] using the Plant Genomic DNA Kit (TianGen Biotech, Beijing, China). The quality and concentration of the extracted DNA were determined by NanoDrop ND 2000 spectrophotometer (NanoDrop Technologies, Inc.) and 1% (w/v) agarose gels electrophoresis. The isolated genomic DNA was diluted to 10 ng/μL by 0.1 × TE buffer (1 mmol/L Tris-HCI, 0.1 mmol/L EDTA, pH = 8.0) and stored at −20 °C.
2.3 Primer selection and SSR–PCR amplification
A total of 102 SSR markers from different resources were used to genotype 64 individuals. Genic-SSR markers with the prefixes LMg [20], LP [25] and B3-B6 [21] were developed from annual ryegrass, perennial ryegrass and Lolium/Festuca complex of grasses, respectively. EST-SSR markers with prefix NFA [23] were developed from tall fescue. All SSR primers (Table 2) were synthesized by Shanghai Sangon Biological Engineering Technology and Services (Shanghai, China).
Polymorphism of 17 SSR markers revealed in 64 individuals of eight Lolium accessions.
| Primer code | SSR repeat motif | Size (bp) | Tm (°C) | N | PIC | Ho |
| LMgSSR02-05D | (AC)14 | 239 | 65.4 | 4 | 0.5188 | 0.7031 |
| LMgSSR03-04F | (AC)11 | 303 | 66.7 | 4 | 0.4172 | 0.2969 |
| LMgSSR03-10B | (AAC)23 | 215 | 66.9 | 6 | 0.5256 | 0.5312 |
| LMgSSR08-10G | (AC)21 | 157 | 66.5 | 7 | 0.6001 | 0.3906 |
| LMgSSR08-12G | (AC)11 | 245 | 67.4 | 5 | 0.5862 | 0.7656 |
| LMgSSR09-09C | (CA)12 | 180 | 68.4 | 5 | 0.7066 | 0.6719 |
| LMgSSR10-10C | (AC)29 | 234 | 64.8 | 6 | 0.4404 | 0.4531 |
| LMgSSR10-12D | (AGG)8(AAG)11(AGG)6 | 266 | 65.6 | 7 | 0.7495 | 0.7188 |
| LMgSSR16-04F | (AC)11 | 242 | 66.8 | 3 | 0.4638 | 0.8594 |
| LMgSSR17-04E | (AC)10 | 179 | 69.0 | 5 | 0.5775 | 0.6875 |
| LPSSRH01H06 | (CA)9 | 150 | 62.2 | 4 | 0.6409 | 0.5625 |
| LPSSRK07H08 | (GAA)7 | 175 | 61.3 | 5 | 0.6755 | 0.5000 |
| NFA012 | (CAG)7 | 203 | 63.5 | 3 | 0.4171 | 0.3281 |
| NFA027 | (TCT)6 | 164 | 63.0 | 5 | 0.4631 | 0.4375 |
| NFA059 | (AG)15 | 170 | 63.7 | 8 | 0.6171 | 0.5469 |
| NFA109 | (CT)10 | 197 | 63.3 | 6 | 0.5983 | 0.6875 |
| B3-B6 | (CA)8 imp. | 238 | 63.6 | 4 | 0.4073 | 0.0781 |
| Min | 3 | 0.4073 | 0.0781 | |||
| Max | 8 | 0.7495 | 0.8594 | |||
| Mean | 5.1176 | 0.5532 | 0.5423 |
Each 15 μL amplification reaction consisted of 3.0 μL of template DNA (10 ng/μL), 0.8 μL primer (5 ng/μL), 0.3 μL of Taq polymerase (2.5 U/μL), 3 μL of sterile distilled water and 7.5 μL of 2 × Taq PCR Master Mix (Tiangen Biotech, Beijing, China). SSRs were amplified under the following PCR conditions: a touchdown PCR consisting of 95 °C for 5 min; 14 cycles of 94 °C for 1 min, (Tm + 2) ∼ (Tm-5)°C for 40 s decreasing by 0.5 °C/cycle and 72 °C for 1 min; and 22 cycles of 95 °C for 40s, (Tm-5)°C for 40 s and 72 °C for 5 min; followed by 72 °C for 5 min and 4 °C as the holding step. PCR amplified fragments were separated using electrophoresis on a 6% denaturing polyacrylamide gel (PAGE) and visualized after silver staining [26].
2.4 Statistical analysis
As a co-dominant nature of SSRs and Lolium species occur as diploid (2n = 2x = 14) [2], allelic bands were scored with genotype based on a molecular DNA marker (50 bp ladder) [27] and formed a “bp” typed original matrix to estimate the number, range and distribution of amplified alleles, and then to determine variation level in the accessions and species. Allele frequencies were computed using GenAlEx 6.5 [28] and polymorphism information content (PIC) was calculated according to Botstein's methods [29].
Supposing the allele frequencies at the species level in accordance with the Hardy–Weinberg equilibrium, POPGENE v. 1.31 [30] was used to estimate Nei's unbiased Genetic Distance (GD) among three species with all markers and assess the following parameters: observed number of alleles (Na), effective number of alleles (Ne), percentage of polymorphic loci (P), Shannon's information index (I), observed heterozygosity (Ho), expected heterozygosity (He), Nei's gene diversity (H), number of polymorphic loci (NPL), the percentage of polymorphic loci (PPL) and the inbreeding coefficient (FIS) according to Weir and Cockerham [31]. The differences between some parameters (PIC, I and He) of markers and species were tested using the ANOVA procedure in the software SPSS version 19. Differences with P values of 0.05 were considered statistically significant. Meanwhile, an analysis of molecular variance (AMOVA) with the program GenAlEx v6.5 was to determine the variance within and among species by ΦPT (analogous to FST) [28] using 9999 random repetitions.
Dendrograms were constructed using an unweighted pair group method of cluster analysis and arithmetic averages (UPGMA) based on the matrix of Nei's GD [32] with POPGENE v. 1.31 [30]. The tree was subsequently visualized by TFPGA v. 1.3 [33] with a bootstrap of 9999 replicates. Also, DataTrans1.0 program [34] was used to transform “bp” typed original data to “0, 1” format for adjusting to NTSYS-pc v2.21r [35] and Principal coordinate analysis (PCoA) was carried out for providing a graphical representation of the genetic relationships between the ryegrass accessions studied by the NTSYS-pc v2.21r [35]. In addition, a Bayesian model-based clustering analysis was used to estimate the most likely number of groups/populations (K) by STRUCTURE 2.3.4 software [36,37], using a burn in length of 50,000, run length of 100,000, and a model allowing for admixture and testing for K from 1 to 9. The results were imported to STRUCTURE HARVESTER software to find the proper number of K by using lnP (D) values [38].
3 Results
3.1 Transferability and polymorphism of SSR markers
From the published reports, a total of 102 SSR primers (gSSRs and eSSRs) previously developed and assayed for L. perenne, L. multiflorum and tall fescue were able to amplify visible DNA fragments for all accessions and no null alleles were found, yielding the overall transferability rate of 100% at the genus level. Most primer pairs produced a complex banding pattern indicating that two or more loci were amplified, and only 17 produced a robust, single-locus pattern with clear signal, good reproducibility, and distinct bands. These 17 single-locus markers were employed in the amplification within 64 individuals, accounting for 16.67%, and 87 alleles were detected, with an average of 5.1 per locus and with correct sizes as expected (Table 2).
In order to evaluate the transferability efficiency, PIC and observed heterozygosity (Ho) for each locus and species are shown in Tables 2 and 3. The PIC value at genus level lied between 0.4073 (B3–B6) and 0.7495 (LMgSSR10-12D), with a mean of 0.5532. According to the classification of PIC [29], of the 17 selected locus, 11 loci demonstrated high polymorphism (PIC ≥ 0.5) and 6 loci demonstrated intermediate polymorphism (0.25 ≤ PIC < 0.5). Ho values varied from 0.0781 (B3-B6) to 0.8594 (LMgSSR16-04F) and averaged at 0.5423. Combined PIC with Ho of each species, comparatively high values were recorded for LMgSSR09-09 C, LMgSSR10-12D and LMgSSR17-04E, thus making them the most polymorphic and informative markers, and revealing a high discriminatory power of the SSR marker system among the genus Lolium.
Mean values of genetic diversity indices calculated over all accessions of L. multiflorum, L. perenne and L. rigidum.
| L. multiflorum | L. perenne | L. rigidum | |
| Number of accessions | 3 | 2 | 3 |
| Number of individuals | 25 | 14 | 25 |
| Number of loci | 17 | 17 | 17 |
| PPL (%) | 100% | 100% | 100% |
| PIC | 0.5139a | 0.4105b | 0.5076a |
| Na | 4.4118 | 3.5882 | 4.1765 |
| Ne | 2.4547 | 1.9750 | 2.5157 |
| I | 1.0600a | 0.8316b | 1.0412a |
| Ho | 0.5812 | 0.3866 | 0.5906 |
| He | 0.5780a | 0.4740b | 0.5795a |
| H | 0.5664 | 0.4571 | 0.5680 |
| FIS | −0.0773 | 0.0828 | −0.1859 |
3.2 Genetic diversity of three Lolium species
Three Lolium species (including eight accessions) comprised of 64 individuals owned a varied genetic diversity reflected by the following parameters including Na, Ne, H, Ho, He, and I when supposing a Hardy–Weinberg equilibrium (HWE). The mean values of genetic diversity indices at the level of each species are presented in Table 3. The percentage of polymorphic loci (PPL) of the genus was 100%. PIC, Na, Ne, H, Ho, He and I were significantly higher in L. multiflorum and L. rigidum than in L. perenne. Besides, fixation indices (FIS, FIT, and FST) are useful tools for studying the genetic differentiation of populations [39]. The mean values of FIS for L. multiflorum, L. perenne and L. rigidum were −0.0773, 0.0828, and −0.1859, respectively.
Table 4 shows the pairwise estimates of ΦPT values between pairs of Lolium accessions. Overall, the mean ΦPT value was about 0.28, indicating that about 28% of the gene diversity was due to differentiation between pairs of accessions. The lowest ΦPT value (0.008) was found between PI 187220 and PI 239486 (L. multiflorum), while the highest (0.542) was between PI 619018 (L. perenne) and PI 545666 (L. rigidum) (Table 4). The AMOVA results suggested that all three levels contributed significantly to the overall genetic variation, with the species level contributing the least (Table 5). Specifically, the three species contributed 17% of the total genetic variance. The next level, between accessions within species, contributed 15% of the total genetic variance, while the remaining 68% genetic variance came from within individual accessions (Table 5). In addition, the total differentiation among populations (ΦPT) was the smallest in L. rigidum (0.29) when compared to L. multiflorum (0.07) and L. perenne (0.18). All components of molecular variance were highly significant (P < 0.001).
Pairwise ΦPT values between Lolium accessions.
| Accessions | LM1 | LM2 | LM3 | LP1 | LP2 | LR1 | LR2 | LR3 |
| PI 187220 (LM1) | 0.000 | |||||||
| PI 239486 (LM2) | 0.008 | 0.000 | ||||||
| PI 241913 (LM3) | 0.090 | 0.120 | 0.000 | |||||
| PI 418725 (LP1) | 0.213 | 0.281 | 0.182 | 0.000 | ||||
| PI 619018 (LP2) | 0.327 | 0.358 | 0.350 | 0.178 | 0.000 | |||
| PI 314447 (LR1) | 0.183 | 0.240 | 0.160 | 0.228 | 0.362 | 0.000 | ||
| PI 422586 (LR2) | 0.291 | 0.336 | 0.267 | 0.396 | 0.449 | 0.224 | 0.000 | |
| PI 545666 (LR3) | 0.323 | 0.401 | 0.373 | 0.361 | 0.542 | 0.252 | 0.379 | 0.000 |
Analysis of molecular variance (AMOVA) of the three Lolium species.
| Source of variation | d.f. | SS | MS | Estimated variance |
% of total variation | Stat | Value | P* |
| Total of three species | ||||||||
| Among species | 2 | 54.608 | 27.304 | 0.856 | 16.74 | Φ RT | 0.1674 | 0.0001 |
| Among accessions within species | 5 | 48.766 | 9.753 | 0.779 | 15.22 | Φ PR | 0.1828 | 0.0001 |
| Within accessions | 56 | 194.891 | 3.480 | 3.480 | 68.04 | Φ PT | 0.3196 | 0.0001 |
| Total | 63 | 298.266 | 5.115 | 100 | ||||
| L. multiflorum | ||||||||
| Among accessions | 2 | 13.148 | 6.574 | 0.315 | 7.38 | Φ PT | 0.0738 | 0.0001 |
| Within accessions | 22 | 86.972 | 3.953 | 3.953 | 92.62 | 0.0001 | ||
| Total | 24 | 100.120 | 4.268 | 100 | ||||
| L. perenne | ||||||||
| Among accessions | 1 | 8.286 | 8.286 | 0.713 | 17.77 | Φ PT | 0.1777 | 0.0001 |
| Within accessions | 12 | 39.571 | 3.298 | 3.298 | 82.23 | 0.0001 | ||
| Total | 13 | 47.857 | 4.010 | 100 | ||||
| L. rigidum | ||||||||
| Among accessions | 2 | 27.333 | 13.666 | 1.269 | 29.00 | Φ PT | 0.2900 | 0.0001 |
| Within accessions | 22 | 68.347 | 3.107 | 3.107 | 71.00 | 0.0001 | ||
| Total | 24 | 95.680 | 4.376 | 100 |
On the basis of Nei's GD, eight accessions could be grouped into three clusters (LM, LP and LR) with a mean threshold of 0.250 as shown on the UPGMA dendrogram (Fig. 1). Also, accessions within species always tended to the same cluster firstly and then to related species, which was similar to the STRUCTURE plot (K = 3). Actually, the model-based simulation of population structure using STRUCTURE software showed that LnP (D) was maximized when K was set at 2 (Fig. 2). It might be explained by higher differentiation in L. multiflorum and L. perenne, with bootstrap values of 90.95 and 91.43, than in L. rigidum (33.61). The relationships observed in the UPGMA analysis and the STRUCTURE analysis were mirrored in the principal coordinate analysis (PCO) (Fig. 3). The first two axes explained only 22.65% of the cumulative molecular variance among accessions, with PCO 1 accounting for 12.92% and PCO 2 for 9.73%.
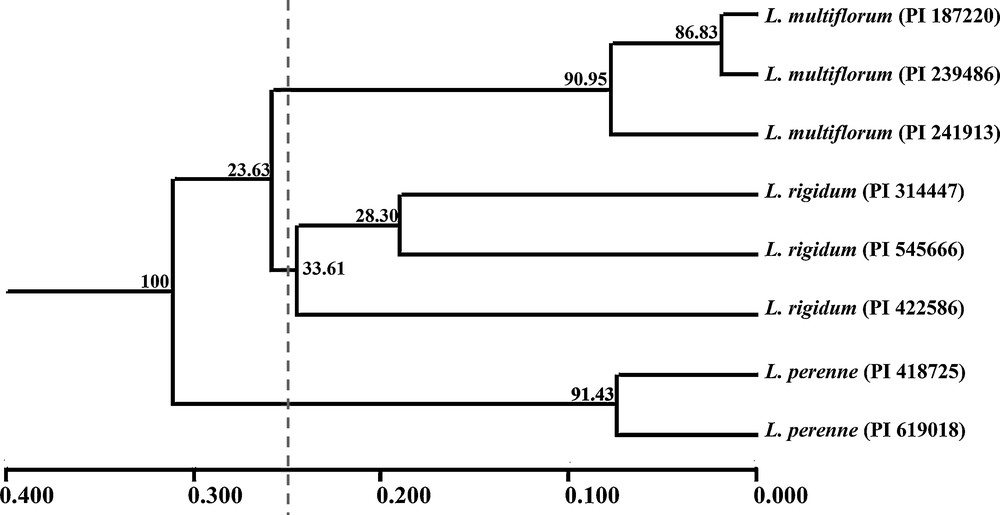
UPGMA dendrogram of eight accessions belonging to L. multiflorum, L. perenne and L. rigidum. Note: The numbers on the branches were verified by bootstrapping analysis to assess the robustness of the dendrogram topology from 9999 replicates.
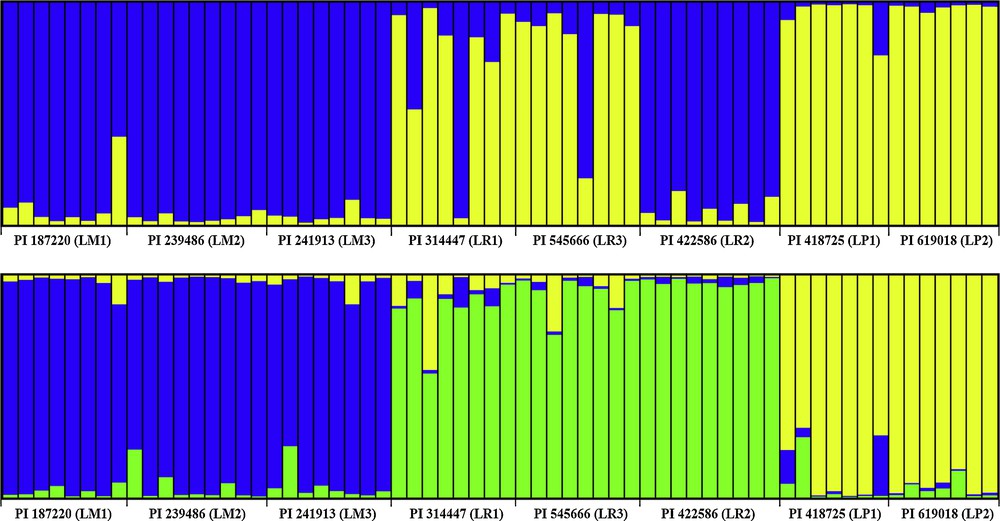
Estimated population structure and genetic admixture among eight accessions belong to L. multiflorum, L. perenne, and L. rigidum using STRUCTURE for K = 2 and 3. Note: Each individual is represented by a single color line. The greater proportion of a color (blue, gold and green), the greater the possibility that the represented individual belongs to the group indicated by that color.
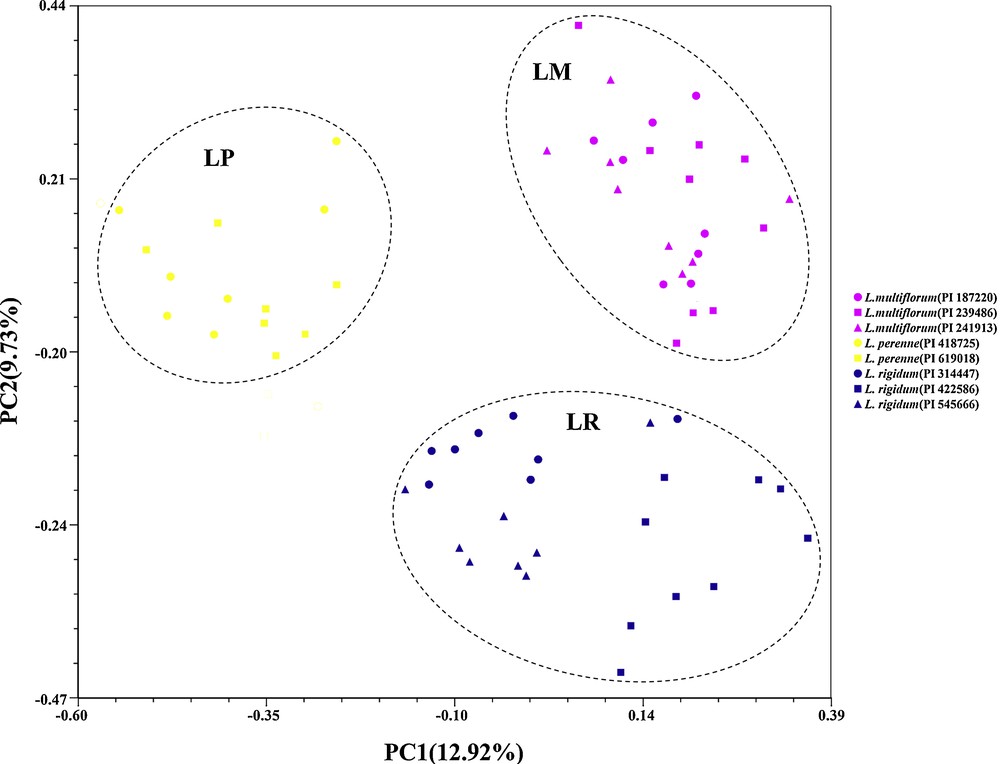
Principal coordinate analysis (PCoA) of 64 individuals wihin eight accessions among L. multiflorum, L. perenne, and L. rigidum. Note: Genetic relationships are depicted by the first two components (PCO1 and PCO2) derived from PCoA of the SSR data. Each species is represented by a single color; pink: L. multiflorum; yellow: L. perenne; and blue: L. rigidum. Different accessions of the same species are indicated by the various symbols (, , ) listed on the right.
4 Discussion
Microsatellites are proved to be conserved in many plant species and could be used in related species [14]. The present studies demonstrate that microsatellite primers developed from Lolium and Festuca species [17,21,23,25] are highly transferable (100%) among the three Lolium species (L. multiflorum, L. perenne, and L. rigidum). This can be explained by a number of causes. First, the relationship between Lolium and broad-leaved fescues are very close and they may own a common ancestor [40,41]. Second, homology locus/regions are well conserved among species of an evolutionary process, even though they belong to the cross-pollinated group, thus they possess some similar alleles [42]. This can result from that there exist Lolium/Festuca complex contact zones where the evolution of introgression might counteract the evolution of isolating mechanisms [43].
However, only 17 single-locus markers were chosen for high polymorphism (the average PIC and Ho value for genus were 0.5532 and 0.5423, respectively) and unambiguous genotypes in the present study; most primers amplified multiple-band patterns similar to dominant markers rather than typical SSR banding patterns. Amplification of multiple loci by some SSR markers might be derived from simultaneous amplification of paralogous loci, which may be due to the recentness of the genome duplication or duplicated genomic regions, as well as speciation [22]. This is in agreement with previous reports on millet [44,45], potato [46], zoysiagrass [47]. Besides, some primer pairs may require different PCR conditions for different samples (species or accessions) to achieve uniform quality of data (i.e. obtaining clear, single-locus phenotypes for some samples may require varying the annealing stringency) [48].
According to AMOVA analysis, most of the total diversity was accounted for by within-populations variation component for all the three species and only 16.74% gene differentiation existed among species, which are in agreement with previous studies [25,49,50]. However, the mean coefficient of gene differentiation is significantly higher in L. rigidum (ΦPT = 0.2900). This is probably because the source of L. rigidum in the present study is collected for a larger area scope (e.g., Georgia, Morocco, and Turkey), where the ecological and climate conditions are diverse, which may result in a greater variety of genetic differentiation.
The analysis of the genetic diversity structure exhibited an organization of accessions which is typically continuous, edifying the relatively close relationships of the three ryegrass species. The PCO analysis based on Dice's similarity distinguish each other among L. multiflorum, L. perenne, and L. rigidum. These results presented sameness with the cluster analysis obtained through UPGMA, confirming the distinctiveness of the three studied species. While comparing differences on the plots (K = 2 and 3) (Fig. 2), L. rigidum showed complex ancestral resource and it revealed that the three studied allogamous ryegrass species might possess some shared alleles for ancestral or ongoing gene flow [51]. Besides, the L. multiflorum accessions are closer to each other with L. rigidum accession than L. perenne, which can be explained by Borrill [52], who suggests that L. multiflorum, also named L. italicum, may have been derived by mutation from an L. rigidum Italian population, which would have been selected by farmers during the thirteenth century. Also, analogous results have already been described via isozyme [40], morphological analysis, and electrophoresis of seed proteins [53,54]. Nevertheless, according to RAPD [55,56] or AFLP data [57], L. multiflorum has a position closer to that of L. perenne than to that of L. rigidum. Several factors might explain the above controversy, such as different clustering methods, characteristics of genetic markers, sample size, and number of studied species.
5 Conclusion
In summary, these findings give an evidence for the potential transferability of SSRs in related Lolium–Festuca species. The relatively high percentage of cross-transferability of these SSR markers to genotypes suggests that these markers are useful for investigating the genetic diversity and structure of the L. rigidum population. The success of cross-amplification suggested that these microsatellite loci can be used for the development of genotype/cultivar specific markers and to explore the genetic relationship among them, as well as for future studies, such as genetic diversity evaluation, gene mapping, and molecular breeding.
Author contributions
Conceived and designed the experiment: Xiao Ma and Xinquan Zhang. Performed the experiment: Zhihui Guo, Kaixin Fu, Chenglin Zhang, Ming Sun and Ting Huang. Analyzed the data: Zhihui Guo, Xiao Ma and Kaixin Fu. Contributed reagents/materials/analysis tools: Xiao Ma, Yan Peng, Yanhong Yan and Linkai Huang. Wrote the paper: Zhihui Guo and Xiao Ma.
Acknowledgments
We thank all the participants from the Department of Grassland Science at Sichuan Agricultural University for their contributions to this study. This work was supported by the National Key Basic Research Program of China (973 Program) (#2014CB138705) and the Earmarked Fund for the Modern Agro-Industry Technology Research System (#CARS-35-05). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.


