1 Introduction
Brassica oleracea L. is one of the most economically important vegetable crop species of the genus Brassica L. in the tribe Brassiceae, which in turn belongs to the family Brassicaceae [1]. This species includes many important cultivars called cole crops [2], comprising cabbage (B. oleracea subspecies capitata), cauliflower (B. oleracea subsp. botrytis), Brussels sprout (B. oleracea subsp. gemmifera), broccoli (B. oleracea subsp. italica), Kale and collards (B. oleracea subsp. acephala), and kohlrabi (B. oleracea subsp. gongylodes). Assessment of genetic diversity, population structure, and relationships is very essential for crop characterisation and conservation, which in turn are important to the continued maintenance and enhancement of agricultural production, leading to sustainable development and global food security [3,4]. Although the phylogeny and genetic diversity of Brassica species have been significantly investigated in the past decade [4–7], there are no reported genetic studies to our knowledge on B. oleracea species in the small island of Ireland [8], whose economy is depending largely on the agricultural production of highly agronomic crops in order to meet the needs of the increasing number of populations. Large numbers of those B. oleracea genetic resources have been collected from different environments and locations throughout Ireland in 1980s, and deposited at the Horticultural Research Institute (HRI) in the United Kingdom. The use of those GenBank resources is being very limited due to their uncompleted characterisation, imprecise phylogeny, and the unmanageable large numbers of accessions that are poor and in danger [8].
Since investigation of the phylogenetic relationships and genetic variation based on morphological and cytological traits could be influenced by environmental factors [9–11], molecular markers have been established and proven to be powerful tools for assessing the genetic diversity and phylogenetic relationships in plants. These molecular markers include AFLP (Amplified Fragment Length Polymorphism) technique developed by Vos et al. [12]. Although it is expensive, AFLP technique proved to be very effective and powerful when compared to other molecular techniques such as Restriction Fragment Length Polymorphism (RFLP) and Random Amplified Polymorphic DNA (RAPD), due to its ability to detect various polymorphisms in various genomic regions that allow the differentiation of closely related species as well as its highly reproducible data and larger numbers of amplified products generated in a single reaction [13]. This method has been successfully used for investigating phylogeny and genetic diversity in Brassica and many other plant species for example, sesame [14], common bean [15], potato [16] and Brassica [4–7,17]. However, those studies did not cover the Irish B. oleracea species. Consequently, the main objective of the current study was to use the powerful AFLP technique to assess the genetic diversity and phylogenetic relationships of a set of the Irish B. oleracea genetic resources deposited at the Horticultural Research Institute (HRI), United Kingdom.
2 Materials and methods
2.1 Plant material
Twenty-five accessions of Irish B. oleracea were obtained from the germplasm collection maintained at the Horticultural Research Institute (HRI), United Kingdom (Table 1). These accessions were chosen based on their sampling site covering a diverse geographical range of the island of Ireland. The selected accessions represented 4 subspecies within B. oleracea species (B. oleracea capitata, B. oleracea acephala, B. oleracea botrytis and B. oleracea gemmifera).
Accession numbers, crop names, and collection sites of the accessions of Brassica oleracea studied.
| No. | Accession Number | Subspecies | Accession name | Crop name | Collection site |
| 1 | HRIGRU 4502 | B. oleracea acephala | Marrow Stem | Fodder kale | Kildare |
| 2 | HRIGRU 4503 | B. oleracea acephala | Thousand Head | Fodder kale | Kildare |
| 3 | HRIGRU 7229 | B. oleracea acephala | Cut and Come Again | Kale | Tipperary |
| 4 | HRIGRU 7556 | B. oleracea acephala | Cut and Come Again | Kale | Cork |
| 5 | HRIGRU 7227 | B. oleracea acephala | Raggedy Jack | Kale | Sligo |
| 6 | HRIGRU 4492 | B. oleracea botrytis | Winter Roscoff | Winter cauliflower | Dublin |
| 7 | HRIGRU 4565 | B. oleracea botrytis | Winter cauliflower | Cork | |
| 8 | HRIGRU 4495 | B. oleracea botrytis | Winter Roscoff | Winter cauliflower | Ballykea |
| 9 | HRIGRU 4579 | B. oleracea capitata | Flat Dutch | Cattle cabbage | Donegal |
| 10 | HRIGRU 4561 | B. oleracea capitata | Flat Dutch | Cattle cabbage | Galway |
| 11 | HRIGRU 4508 | B. oleracea capitata | Flat Dutch | Cattle cabbage | Ballina |
| 12 | HRIGRU 4506 | B. oleracea capitata | Flat Dutch | Cattle cabbage | Ballinrobe |
| 13 | HRIGRU 4585 | B. oleracea capitata | Flat Dutch | Common cabbage | Donegal |
| 14 | HRIGRU 4586 | B. oleracea capitata | Flat Dutch | Common cabbage | Mayo |
| 15 | HRIGRU 4497 | B. oleracea capitata | Flat Dutch | Cabbage | Roscommon |
| 16 | HRIGRU 4498 | B. oleracea capitata | Flat Dutch | Cabbage | Roscommon |
| 17 | HRIGRU 4588 | B. oleracea capitata | Flat Dutch | Cabbage | Donegal |
| 18 | HRIGRU 5915 | B. oleracea capitata | Flat Dutch | Cabbage | Limerick |
| 19 | HRIGRU12532 | B. oleracea capitata | Delaway Cabbage | Cabbage | Mayo |
| 20 | HRIGRU 4566 | B. oleracea capitata | Spring cabbage | Cork | |
| 21 | HRIGRU 4564 | B. oleracea capitata | Spring cabbage | Cork | |
| 22 | HRIGRU 4571 | B. oleracea capitata | Spring cabbage | Cork | |
| 23 | HRIGRU 5914 | B. oleracea capitata | Spring Greens | Spring cabbage | Limerick |
| 24 | HRIGRU 4491 | B. oleracea gemmifera | Brussels sprout | Dublin | |
| 25 | HRIGRU 4494 | B. oleracea gemmifera | Brussels sprout | Dublin |
2.2 DNA extraction
Genomic DNA was isolated from 3-week old leaf tissue using DNeasy Plant Mini Kit (Qiagen, United Kingdom), following the procedures described by manufacturers. Five DNA samples were prepared from each of the 25 accessions studied and were subjected to AFLP analysis.
2.3 AFLP analysis
Sixteen AFLP primer sets were selected from the literature [18,19] and used in a screening test for polymorphisms using 2 different cabbage accessions. Following the screening, 11 primer sets revealed the highest polymorphism and used to analyse all the accessions (Table 2).
AFLP primer sets, number and size of AFLP fragments amplified and Nei's [21] genetic diversity indices in the 25 accessions of Brassica oleracea studied.
| Primer sets | EcoRI-AAC/MseI-CAG | EcoRI-AGG/MseI-CTA | EcoRI-AAG/MseI-CAA | EcoRI-ACT/MseI-CAA | EcoRI-AGG/MseI-CAG | EcoRI-AGG/MseI-CAA | EcoRI-ACT/MseI-CAG | EcoRI-ACT/MseI-CTA | EcoRI-AAC/MseI-CTA | EcoRI-AAC/MseI-CAA | EcoRI-AAG/MseI-CTA | Mean ± Standard Deviation |
| Total number of fragments | 37 | 49 | 40 | 45 | 43 | 44 | 43 | 44 | 43 | 41 | 42 | 42.82 ± 3.02 |
| Number of polymorphic fragments | 37 | 40 | 38 | 38 | 39 | 38 | 39 | 39 | 38 | 38 | 39 | 38.45 ± 0.82 |
| Percentage of polymorphic loci (%) | 100 | 81.6 | 95 | 84.4 | 90.7 | 86.4 | 90.7 | 88.5 | 88.4 | 92.7 | 92.9 | 89.8 |
| Lowest molecular size (bp) | 46 | 40 | 48 | 44 | 41 | 43 | 42 | 47 | 45 | 44 | 46 | 44.18 ± 2.52 |
| Highest molecular size (bp) | 992 | 997 | 960 | 890 | 900 | 930 | 900 | 930 | 945 | 900 | 930 | 934 ± 36.85 |
| A e | 1.470 | 1.414 | 1.438 | 1.423 | 1.423 | 1.438 | 1.423 | 1.425 | 1.424 | 1.427 | 1.426 | 1.43 ± 0.015 |
| H T | 0.262 | 0.239 | 0.256 | 0.245 | 0.241 | 0.245 | 0.249 | 0.242 | 0.261 | 0.259 | 0.260 | 0.251 ± 0.009 |
| H S | 0.187 | 0.139 | 0.159 | 0.169 | 0.141 | 0.143 | 0.147 | 0.149 | 0.156 | 0.164 | 0.161 | 0.156 ± 0.014 |
| D ST | 0.075 | 0.100 | 0.097 | 0.076 | 0.081 | 0.086 | 0.087 | 0.092 | 0.095 | 0.091 | 0.089 | 0.088 ± 0.008 |
| G ST | 0.286 | 0.418 | 0.379 | 0.310 | 0.390 | 0.299 | 0.355 | 0.376 | 0.289 | 0.291 | 0.312 | 0.337 ± 0.048 |
AFLP analysis was carried out using a modified protocol described by Vos et al. [12]. In brief, the restriction-ligation reactions were prepared in a final volume of 20 μl, containing 1 μl of DNA sample (100 ng/μl), 1 μl of enzyme master mixture (Medical Supply Co. Ltd), 2 μl of T4 ligase buffer (with ATP), 1 μl EcoRI adaptor (Applied Biosystems), 1 μl of MseI adaptor (Applied Biosystems) and 14 μl of molecular grade water. These restriction-ligation mixtures were incubated at 37 °C for 2 hours and then denatured at 70 °C for 15 minutes. The preselective amplification reactions were prepared in a final volume of 25 μl containing 3 μl of the diluted restriction-ligation DNA preparation, 1.25 μl of each of the preselective primers (50 ng/μl) (Medical Supply Co. Ltd), 12.5 μl of GoTaq® green master mixture (Medical Supply Co. Ltd) and 7 μl of water. The pre-selective PCR reactions were then amplified in a DNA thermocycler programmed under the following conditions: 94 °C for 2 min (1 cycle); 94 °C for 1 min, 56 °C for 1 min, 72 °C for 1 min (26 cycles) and 72 °C for 30 min (1 cycle). The preselective amplification products were then visualised on a 1.5% agarose gel. The selective amplification reactions were prepared in a final volume of 25 μl containing 2 μl of template DNA from pre-selective PCR, 2 μl of selective primer set (50 ng/μl) (Eurofins MWG Operon), 12.5 μl of GoTaq® green master mixture and 8.5 μl of water (nuclease free). The selective PCR reactions were then amplified in a DNA thermocycler programmed as follows: 1 cycle at 94 °C for 30 s, 65 °C for 30 s and 72 °C for 60 s. The annealing temperature was then lowered by 0.7 °C per cycle during the first 12 cycles, and then 23 cycles were performed at 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 60 s, then at 72 °C for 5 min. The amplification products were mixed with loading solution and then separated by 8% (w/v) polyacrylamide gel electrophoresis. 50 bp DNA ladder was used as a DNA standard (Medical Supply Co. Ltd). Gels were stained with SYBR Gold solution (250 ml of 1X TAE and 25 μl of SYBR Gold molecular stain), and photographed.
2.4 Data analysis
AFLP data were analysed using GelCompar II version 6.0 Applied Maths, SYSTAT for Windows version 7.0, SPSS INC, and POPGENE version 1.31 software. AFLP bands were visually scored as present (1) and absent (0) to create the binary data set. The dendrogram was constructed based on Nei's genetic distance using unweighted pair group method with arithmetic average (UPGMA) [20]. The partitioning of total genetic diversity into within- and among-accession components was examined using Nei's [21] genetic diversity statistics. The partitioning of total genetic diversity (HT) into within- (HS) and among- (DST) accession components was examined using Nei's [20,21] genetic diversity statistics (HT = HS + DST; GST = DST/HT).
3 Results
3.1 Polymorphism analysis of AFLP data
A total of 471 fragments were scored across the 11 AFLP primer pairs assayed in this study, out of which 423 (89.8%) were polymorphic (Table 2). The fragments generated ranged in size from 40 to 997 bp. The number of scorable fragments amplified by each primer pair varied from 37 to 49 with an average of 42.82 (Table 2). The primer pair (EcoRI-AAC/MseI-CAG) amplified the lowest number of fragments (37), with sizes ranging from 46 to 992 bp. The primer pair (EcoRI-AGG/MseI-CTA) amplified the highest number of fragments (49), with sizes ranging from 40 to 997 bp. The number of polymorphic fragments for each primer pair also varied from 37 to 40 with an average of 38.45. However, the percentage of polymorphic loci ranged from 81.6 to 100% with an average of 89.8% (Table 2).
3.2 Genetic structure and Nei's diversity indices
Genetic variation in accessions was measured in terms of the effective number of alleles and the mean genetic diversity (Table 2). The averages of total genetic diversity in the accessions studied (HT) and intra-accessional genetic diversity (HS) were 0.251 and 0.156, respectively (Table 2). Moreover, the inter-accessional genetic diversity (DST) varied from 0.075 to 0.1 with an average of 0.088, and the coefficient of genetic differentiation among accessions (GST) varied from 0.286 to 0.418 with an average of 0.337. The effective number of alleles (Ae) varied from 1.414 to 1.47, with a mean of 1.43.
3.3 Similarity indices and genetic distance
The overall mean similarity index for B. oleracea accessions calculated based on all AFLP fragments amplified using Nei's [20] similarity index, ranged from 0.297 to 0.999 with an average of 0.744 (Table 3). The highest similarity indices (0.999) and the lowest genetic distance (0.001) were between the accessions of the same crop variety and geographical region, e.g., spring cabbage HRIGRU 4564 and HRIGRU 4571 from Cork. Accessions having close proximity in their origin, breeding strategies and morphological traits are likely to have less genetic distance from each other. The second highest similarity indices (0.998) were between the accessions of cabbages HRIGRU 4497 from Roscommon and HRIGRU 4564 from Mayo. While the highest genetic distance (0.703) and the lowest similarity indices (0.297) were found between the accessions of brussels sprout HRIGRU 4494 and kale HRIGRU 7556. The highest genetic distance in the cabbage types was found between the accessions of cabbage HRIGRU 4588 and cattle cabbage HRIGRU 4561 and HRIGRU 4508.
Sj similarity indices (above diagonal) and pair wise genetic distance values (below diagonal) calculated from AFLP data of the 25 accessions of Brassica oleracea (see Table 1 for identification of accessions numbers).
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | |
| 1 | **** | 0.954 | 0.953 | 0.944 | 0.948 | 0.694 | 0.635 | 0.665 | 0.531 | 0.502 | 0.523 | 0.504 | 0.515 | 0.519 | 0.522 | 0.500 | 0.539 | 0.538 | 0.516 | 0.498 | 0.545 | 0.547 | 0.501 | 0.51 | 0.343 |
| 2 | 0.047 | **** | 0.977 | 0.956 | 0.928 | 0.678 | 0.608 | 0.638 | 0.553 | 0.524 | 0.545 | 0.520 | 0.536 | 0.54 | 0.543 | 0.521 | 0.560 | 0.554 | 0.532 | 0.508 | 0.566 | 0.568 | 0.523 | 0.534 | 0.35 |
| 3 | 0.048 | 0.024 | **** | 0.987 | 0.959 | 0.664 | 0.624 | 0.654 | 0.556 | 0.527 | 0.548 | 0.523 | 0.539 | 0.543 | 0.546 | 0.525 | 0.563 | 0.557 | 0.536 | 0.511 | 0.569 | 0.572 | 0.526 | 0.515 | 0.313 |
| 4 | 0.058 | 0.045 | 0.013 | **** | 0.949 | 0.651 | 0.615 | 0.645 | 0.536 | 0.508 | 0.529 | 0.504 | 0.520 | 0.524 | 0.527 | 0.506 | 0.544 | 0.538 | 0.517 | 0.493 | 0.549 | 0.552 | 0.507 | 0.507 | 0.297 |
| 5 | 0.053 | 0.075 | 0.042 | 0.052 | **** | 0.675 | 0.639 | 0.669 | 0.547 | 0.519 | 0.539 | 0.519 | 0.531 | 0.535 | 0.538 | 0.516 | 0.555 | 0.558 | 0.532 | 0.514 | 0.560 | 0.563 | 0.531 | 0.512 | 0.343 |
| 6 | 0.365 | 0.388 | 0.409 | 0.429 | 0.394 | **** | 0.917 | 0.888 | 0.795 | 0.763 | 0.774 | 0.749 | 0.792 | 0.772 | 0.794 | 0.781 | 0.835 | 0.805 | 0.783 | 0.783 | 0.827 | 0.819 | 0.784 | 0.549 | 0.422 |
| 7 | 0.455 | 0.497 | 0.472 | 0.486 | 0.448 | 0.086 | **** | 0.959 | 0.797 | 0.781 | 0.769 | 0.769 | 0.812 | 0.783 | 0.797 | 0.795 | 0.836 | 0.808 | 0.786 | 0.791 | 0.834 | 0.827 | 0.780 | 0.529 | 0.385 |
| 8 | 0.408 | 0.449 | 0.425 | 0.438 | 0.402 | 0.119 | 0.041 | **** | 0.753 | 0.744 | 0.748 | 0.757 | 0.796 | 0.769 | 0.777 | 0.783 | 0.793 | 0.764 | 0.766 | 0.775 | 0.794 | 0.787 | 0.779 | 0.489 | 0.309 |
| 9 | 0.633 | 0.593 | 0.588 | 0.623 | 0.603 | 0.230 | 0.227 | 0.284 | **** | 0.983 | 0.966 | 0.969 | 0.965 | 0.955 | 0.972 | 0.964 | 0.959 | 0.948 | 0.972 | 0.957 | 0.962 | 0.960 | 0.947 | 0.658 | 0.583 |
| 10 | 0.688 | 0.647 | 0.640 | 0.677 | 0.656 | 0.271 | 0.247 | 0.295 | 0.017 | **** | 0.95 | 0.983 | 0.949 | 0.961 | 0.946 | 0.961 | 0.929 | 0.922 | 0.946 | 0.938 | 0.945 | 0.945 | 0.939 | 0.666 | 0.596 |
| 11 | 0.648 | 0.607 | 0.601 | 0.637 | 0.617 | 0.256 | 0.263 | 0.291 | 0.034 | 0.052 | **** | 0.973 | 0.954 | 0.947 | 0.958 | 0.944 | 0.923 | 0.914 | 0.953 | 0.934 | 0.928 | 0.927 | 0.944 | 0.629 | 0.528 |
| 12 | 0.686 | 0.654 | 0.647 | 0.685 | 0.654 | 0.288 | 0.262 | 0.278 | 0.031 | 0.017 | 0.027 | **** | 0.961 | 0.957 | 0.955 | 0.971 | 0.918 | 0.911 | 0.960 | 0.962 | 0.931 | 0.929 | 0.952 | 0.655 | 0.571 |
| 13 | 0.664 | 0.624 | 0.618 | 0.653 | 0.633 | 0.234 | 0.208 | 0.228 | 0.036 | 0.052 | 0.048 | 0.039 | **** | 0.974 | 0.969 | 0.967 | 0.961 | 0.934 | 0.959 | 0.964 | 0.959 | 0.952 | 0.959 | 0.635 | 0.525 |
| 14 | 0.656 | 0.616 | 0.610 | 0.646 | 0.626 | 0.259 | 0.244 | 0.262 | 0.046 | 0.039 | 0.055 | 0.044 | 0.026 | **** | 0.969 | 0.965 | 0.936 | 0.933 | 0.958 | 0.942 | 0.953 | 0.954 | 0.938 | 0.641 | 0.534 |
| 15 | 0.650 | 0.610 | 0.605 | 0.639 | 0.619 | 0.230 | 0.227 | 0.252 | 0.028 | 0.055 | 0.043 | 0.046 | 0.031 | 0.032 | **** | 0.988 | 0.964 | 0.961 | 0.998 | 0.975 | 0.972 | 0.974 | 0.960 | 0.655 | 0.569 |
| 16 | 0.693 | 0.651 | 0.645 | 0.682 | 0.661 | 0.247 | 0.230 | 0.245 | 0.037 | 0.039 | 0.058 | 0.029 | 0.033 | 0.035 | 0.013 | **** | 0.951 | 0.939 | 0.988 | 0.983 | 0.964 | 0.962 | 0.960 | 0.661 | 0.588 |
| 17 | 0.618 | 0.580 | 0.574 | 0.609 | 0.589 | 0.181 | 0.179 | 0.232 | 0.042 | 0.073 | 0.080 | 0.086 | 0.039 | 0.066 | 0.037 | 0.051 | **** | 0.976 | 0.953 | 0.953 | 0.988 | 0.989 | 0.949 | 0.666 | 0.573 |
| 18 | 0.621 | 0.590 | 0.585 | 0.619 | 0.583 | 0.216 | 0.214 | 0.269 | 0.054 | 0.082 | 0.089 | 0.093 | 0.068 | 0.069 | 0.039 | 0.063 | 0.024 | **** | 0.955 | 0.937 | 0.985 | 0.986 | 0.924 | 0.670 | 0.572 |
| 19 | 0.662 | 0.630 | 0.624 | 0.660 | 0.631 | 0.244 | 0.241 | 0.266 | 0.028 | 0.055 | 0.048 | 0.041 | 0.042 | 0.043 | 0.002 | 0.013 | 0.048 | 0.046 | **** | 0.985 | 0.961 | 0.963 | 0.952 | 0.653 | 0.569 |
| 20 | 0.697 | 0.676 | 0.670 | 0.503 | 0.665 | 0.244 | 0.235 | 0.255 | 0.044 | 0.064 | 0.068 | 0.039 | 0.037 | 0.059 | 0.026 | 0.017 | 0.048 | 0.065 | 0.015 | **** | 0.951 | 0.943 | 0.957 | 0.645 | 0.548 |
| 21 | 0.608 | 0.569 | 0.564 | 0.598 | 0.579 | 0.190 | 0.182 | 0.231 | 0.039 | 0.056 | 0.075 | 0.071 | 0.042 | 0.048 | 0.028 | 0.037 | 0.004 | 0.015 | 0.039 | 0.05 | **** | 0.999 | 0.946 | 0.684 | 0.599 |
| 22 | 0.603 | 0.565 | 0.559 | 0.594 | 0.574 | 0.199 | 0.190 | 0.239 | 0.041 | 0.057 | 0.076 | 0.073 | 0.049 | 0.047 | 0.027 | 0.039 | 0.011 | 0.014 | 0.038 | 0.058 | 0.001 | **** | 0.939 | 0.686 | 0.603 |
| 23 | 0.690 | 0.649 | 0.643 | 0.679 | 0.634 | 0.243 | 0.247 | 0.249 | 0.054 | 0.063 | 0.058 | 0.049 | 0.042 | 0.064 | 0.041 | 0.041 | 0.053 | 0.079 | 0.049 | 0.044 | 0.055 | 0.063 | **** | 0.655 | 0.562 |
| 24 | 0.669 | 0.626 | 0.663 | 0.679 | 0.669 | 0.598 | 0.635 | 0.511 | 0.419 | 0.406 | 0.463 | 0.424 | 0.454 | 0.444 | 0.423 | 0.414 | 0.405 | 0.400 | 0.426 | 0.437 | 0.379 | 0.376 | 0.422 | **** | 0.97 |
| 25 | 0.657 | 0.650 | 0.687 | 0.703 | 0.657 | 0.578 | 0.615 | 0.691 | 0.417 | 0.404 | 0.472 | 0.429 | 0.475 | 0.466 | 0.431 | 0.412 | 0.427 | 0.428 | 0.431 | 0.452 | 0.401 | 0.397 | 0.438 | 0.03 | **** |
3.4 Cluster analysis
The dendrogram constructed based on Nei's [20] genetic distance, showed that the total genetic distance among the accessions examined was 66% (Fig. 1). The dendrogram also showed four major groups. The first group contained all accessions of kale and fodder kale. The second group contained the two accessions of Brussels sprout. The third group was homogenous and contained all accessions of winter cauliflower. The fourth group split into four subgroups, and contained all the accessions of cabbage types. The cattle cabbage and common cabbage formed distinct clusters, whereas the cabbage and spring cabbage were distributed among different clusters within the first and third subgroups. The 4 accessions of cattle cabbage were separated in the second subgroup, and the 2 accessions of common cabbage were separated in one cluster within the fourth subgroup. The cluster analysis also showed that the cauliflowers and cabbages were more closely related to each other.
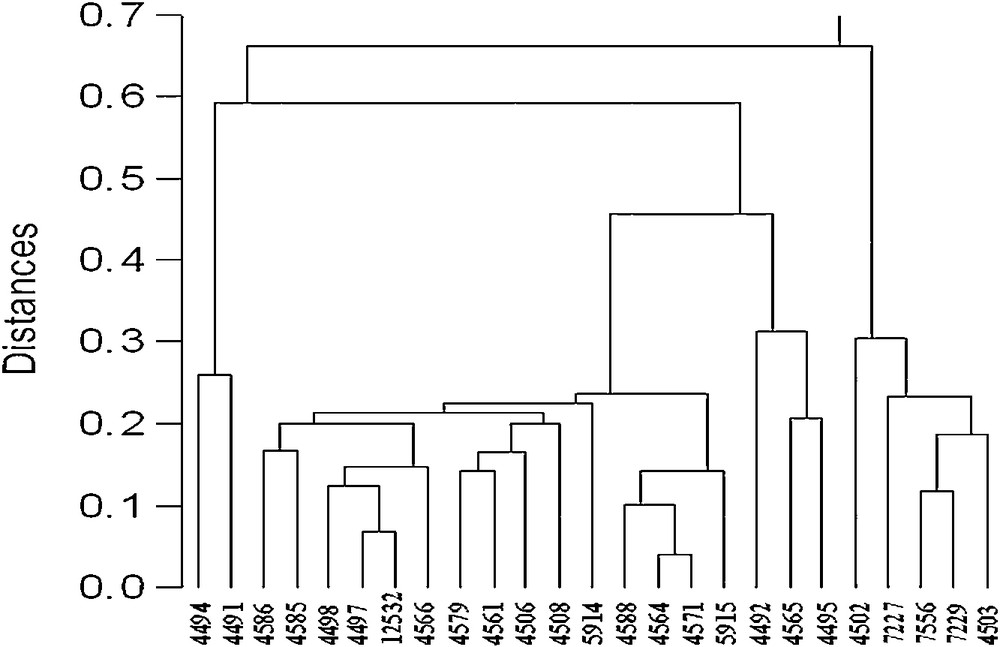
UPGMA dendrogram based on Nei's [20] genetic distance, showing the relationships among 25 accessions of Brassica oleracea based on AFLP data.
3.5 Principal component analysis
Principal component analysis (PCA) showed a pronounced genetic variation among the 25 accessions of B. oleracea studied. The first 4 variables of the principal component analysis accounted for 91% of the total variation in AFLP data generated. The first component (PC1) explained 59.7% of the total variation, whereas the second (PC2), the third (PC3) and the fourth (PC4) components contributed 18.93%, 7.77% and 4.6% of the total variation, respectively (Table 4).
Matrix of eigenvalues of the principal components analysis for the 25 accessions of Brassica oleracea studied based on AFLP data.
| Principal Components | ||||
| PC1 | PC2 | PC3 | PC4 | |
| Variance Explained by Components | 14.925 | 4.731 | 1.943 | 1.150 |
| Percent of Total Variance Explained | 59.698 | 18.925 | 7.773 | 4.599 |
| Accumulated Eigenvectors | 59.698 | 78.623 | 86.396 | 91 |
| HRIGRU 4502Fodder kale | 0.067 | −0.884 | 0.104 | −0.030 |
| HRIGRU 4503Fodder kale | 0.085 | −0.932 | 0.131 | 0.134 |
| Kale HRIGRU 7229 | 0.092 | −0.958 | 0.096 | 0.171 |
| Kale HRIGRU 7556 | 0.050 | −0.946 | 0.089 | 0.163 |
| Kale HRIGRU 7227 | 0.085 | −0.935 | 0.080 | 0.086 |
| Cauliflower HRIGRU 4492 | 0.644 | −0.354 | −0.322 | −0.430 |
| Cauliflower HRIGRU 4565 | 0.672 | −0.263 | −0.417 | −0.496 |
| Cauliflower HRIGRU 4495 | 0.605 | −0.306 | −0.497 | −0.420 |
| Cattle cabbage HRIGRU 4579 | 0.959 | 0.062 | 0.049 | 0.125 |
| Cattle cabbage HRIGRU 4561 | 0.927 | 0.127 | 0.068 | 0.122 |
| Cattle cabbage HRIGRU 4508 | 0.906 | 0.092 | −0.011 | 0.210 |
| Cattle cabbage HRIGRU 4506 | 0.927 | 0.135 | 0.036 | 0.171 |
| Common cabbage HRIGRU 4585 | 0.955 | 0.063 | −0.051 | 0.084 |
| Common cabbage HRIGRU 4586 | 0.937 | 0.067 | −0.002 | 0.140 |
| Cabbage HRIGRU 4497 | 0.972 | 0.057 | 0.015 | 0.096 |
| Cabbage HRIGRU 4498 | 0.964 | 0.104 | 0.022 | 0.071 |
| Cabbage HRIGRU 4588 | 0.956 | −0.004 | −0.004 | −0.053 |
| Cabbage HRIGRU 5915 | 0.934 | 0 | 0.031 | −0.007 |
| Cabbage HRIGRU 12532 | 0.961 | 0.081 | 0.022 | 0.113 |
| Spring cabbage HRIGRU 4566 | 0.943 | 0.124 | −0.020 | 0.076 |
| Spring cabbage HRIGRU 4564 | 0.969 | −0.009 | 0.032 | −0.044 |
| Spring cabbage HRIGRU 4571 | 0.965 | −0.018 | 0.048 | −0.032 |
| Spring cabbage HRIGRU 5914 | 0.930 | 0.092 | 0.003 | 0.066 |
| Brussels sprout HRIGRU 4491 | 0.308 | 0.069 | 0.834 | −0.359 |
| Brussels sprout HRIGRU 4494 | 0.331 | 0.022 | 0.809 | −0.397 |
Table 4 shows the matrix of eigenvalues of the principal component analysis for all accessions of B. oleracea studied. All the accessions of cauliflowers and cabbages studied showed the highest variability on the first component. The 2 accessions of brussels sprouts and the accession of fodder kale HRIGRU 4502 showed their greatest variability on the third component, while all other accessions of kales showed an intermediate level of variation on the fourth component (Table 4).
4 Discussion
The use of molecular markers to assess the genetic diversity and relationships among individuals and populations is very beneficial as many polymorphic loci can be obtained in a relatively short time and at low cost, without any prior knowledge of the genome of the species under study [22,23]. AFLP has emerged as a powerful technique for cultivar identification and fingerprinting [4,6,7,24]. This is an important and comprehensive study that investigates the genetic diversity, population structure and relationships of 25 accessions of the Irish B. oleracea species using the powerful amplified fragment length polymorphism (AFLP) technique. The results of the AFLP validation study showed different banding patterns for the accessions studied. Therefore, AFLP markers could detect multiple polymorphisms among different accessions of B. oleracea, and also proved to be accurate and highly reproducible.
AFLP fragments generated by the 11 AFLP primer pairs assayed in this study were different in number, intensity and position, indicating a high genetic variation of the accessions studied. A total of 471 AFLP fragments were scored across these 11 AFLP primer pairs, of which 423 (89.8%) were polymorphic and could differentiate the accessions analysed, reflecting a rich allelic diversity in the cultivars. This percentage of polymorphic loci (89.8%) was higher than that reported by Lázaro and Aguinagalde [25] and Faltusová et al. [4] for B. oleracea (54% and 45%, respectively), and Das et al. [26] for Brassica campestris (66.8%).
In the current study, the average number of scorable fragments amplified per AFLP primer pair was 42.82, which was higher than that reported by Huh and Huh [27] for B. campestris (42.6) and Genet et al. [17] for Brassica carinata (32). Furthermore, the average number of polymorphic fragments per primer pair was 38.45, and this value was higher than that reported by Yu et al. [28] for Brassica napus (13.4). The effective number of alleles (Ae) also varied from 1.414 to 1.47 with a mean of 1.43, which was lower than that reported by Huh and Huh [27] for B. campestris (3.2). The difference in all of this data could be attributed to the differences in Brassica species or the AFLP primer sets used. The AFLP primers used in this study showed a high degree of polymorphism and are recommended for inclusion in future studies of diversity assessment of Brassica spp.
The cluster analysis of the AFLP data showed that the cauliflowers and cabbages were more closely related to each other. They shared most of the AFLP patterns. The accessions of some cabbage types were distributed among different clusters within cabbage subgroups due to the differences in their AFLP data. These results were in agreement with a previous morphological study [29,30] and with that reported by Balkaya et al. [31]. It is recommended that further comprehensive analysis would be needed for a rigorous comparison to be made. Furthermore, cluster analysis revealed that the accession of cabbage HRIGRU 12532 was grouped with the accessions of cabbages HRIGRU 4497 and HRIGRU 4498, but this result was not in agreement with the morphological data [30], which showed that the accession of cabbage HRIGRU 12532 was not grouped with the same clusters of cabbages. This difference in data could be attributed to the fact that the morphological traits may be influenced by many factors including environmental conditions, sample size and the time of taking measurements.
The overall mean similarity index calculated based on all AFLP fragments amplified using Nei's [20] similarity index, ranged from 0.297 to 0.999 with an average of 0.744, indicating a wide genetic base. This study also suggests a new insight, indicating that the accessions of kale HRIGRU 7556, cattle cabbage HRIGRU 4561 and HRIGRU 4508 and cabbage HRIGRU 4588, due to the highest genetic variation found among them, could provide parental sources in future breeding program to develop new or more productive Brassica varieties. Furthermore, PCA showed different levels of genetic variation among the accessions studied. These differences could be attributed to the breeding system and geographical factors of these accessions. This analysis also showed that 91% of the total variation in our AFLP data was separated on the first 4 components. All the accessions of cauliflowers and cabbages studied showed the highest variability on the first component, indicating a very high degree of correlation among these accessions.
The present study showed that the accessions had a pronounced genetic diversity, but the distribution of this diversity was not homogeneous. The mean value of the total genetic diversity (HT) was 0.251. This value was similar to that reported by Hintum et al. [5] and Watson-Jones et al. [32] for B. oleracea (0.249 and 0.25, respectively), but lower than that reported by Lázaro and Aguinagalde [25,33] for wild taxa of B. oleracea (0.338 and 0.294, respectively). The intra-accessional genetic diversity (HS) was 0.156, whereas this value was higher than that reported by Hintum et al. [5] for B. oleracea (0.13). The differences in these data from various studies on B. oleracea could be attributed to the differences in accessions or the methodology used.
The coefficient of genetic differentiation among accessions in this study indicated that 33.7% of the total genetic variation was due to differences among accessions, and 66.3% of the AFLP variation resided within accessions. Therefore, the majority of the total genetic variation resided within accessions. This result agreed with that reported by Lázaro and Aguinagalde [25], Watson-Jones et al. [32], Hintum et al. [5] and Christensen et al. [7] who all stated that the percentage of genetic variation within B. oleracea accessions was higher than that resided among accessions. The combinations of an insect-pollinated, outcrossing breeding system, a high degree of gene flow and a propensity for high fecundity might explain the high level of genetic diversity within the accessions studied. The AFLP marker system used in this study proved efficient for assessing the genetic diversity and relationships in Irish B. oleracea, and will therefore have potential in the future to distinguish varieties for commercial purposes. Furthermore, using breeding experiments, molecular markers can be utilised for marker-aided selection and quantitative trait loci analysis [34–43].
5 Conclusions
This study assesses the genetic diversity and phylogenetic relationships of the Irish B. oleracea species using the powerful AFLP technique. A total of 423 AFLP fragments were polymorphic and differentiated most of the accessions analysed. The majority of genetic variation resided within accessions. This high level of variability demonstrates that the Irish B. oleracea genetic resources could be managed and conserved for future utilisation and exploitation in food and agriculture. Moreover, this study addressed important phylogenetic questions within this species, and recommended 4 accessions of cabbages and kales for use in future breeding programs, genomic studies and varieties improvement. This phylogenetic analysis may also provide the starting point for further research towards a complete phylogeny for brassicas.
Authors’ contributions
ME designed and performed the experiments, analysed the data and wrote the manuscript. KG contributed to experimental design, analysis and writing of the manuscript. PB contributed to experimental design and writing of the manuscript. RM contributed to the experimental design and to the writing of the manuscript. All authors read and approved the final version of the manuscript.
Disclosure of interest
The authors declare that they have no competing interest.
Acknowledgments
We would like to thank the Horticultural Research Institute at University of Warwick in United Kingdom for providing us with the Irish Brassica material used in this study. We thank Barry Murphy (Teagasc Research Centre, Ireland) for his support during selecting Brassica accessions. We are grateful for the support from Brassica working group in Europe. This work was financially supported by the Department of Agriculture, Fisheries and Food (DAFF, Ireland) under the Conservation of Genetic Resources for Food and Agriculture Scheme 2009 and the Dublin Institute of Technology ABBEST Scholarship Scheme, Ireland.


