1 Introduction
Eurytemora affinis (Poppe, 1880) is a widely distributed species inhabiting fresh- and brackish waters of the Palearctic region. This species is one of the more challenging calanoid copepods for taxonomic identification, due to the absence of highly distinctive sexually dimorphic appendages, high morphological variability and a broad distribution. Because the species is relatively well studied in Europe [1–8], it is important to provide a detailed analysis of the intraspecific structure of E. affinis, using morphological and genetic tools. This holds particularly since some invasions of the E. cf. affinis North-American clade (now Eurytemora carolleeae) in the Baltic Sea were observed [4,8].
Since the first description of E. affinis (Poppe) in 1880, a number of species and subspecies were described within the species: E. affinis hirundo Giesbrecht, 1881, E. affinis raboti Richard, 1897, E. affinis hirundoides Nordquist, 1888, E. affinis hispida (Nordquist, 1888), Eurytemora inermis (Boeck, 1864). Afterwards, some subspecies were accepted as valid species: E. hirundo Giesbrecht, 1881, E. raboti Richard, 1897, E. hirundoides (Nordquist, 1888). Detailed morphometric studies have indicated that E. affinis, E. hirundo (Giesbrecht, 1881), and the more slender E. hirundoides (Nordquist, 1888) are morphological variants of the same species [9–11]. No genetic differences between E. affinis and E. hirundoides were found [12]. Hence, Eurytemora taxonomy turns out to be problematic, in particular because the use of species names is inconsistent among studies with the use of different taxonomic names, depending on different preferences in systematics or identification keys [13,14].
In the last decades, the number of publications on the comparison of E. cf. affinis clades increased, including phylogenetic, population genetic and eco-physiological aspects [4,6,15,16] as well as morphological descriptions [4,8,17–19]. However, there is still no clear allocation of the type species for E. affinis and the type specimen cannot be retrieved.
DNA sequencing offers new opportunities for taxonomic and phylogeographic studies in the past three decades. The analysis of extensive population samples of E. cf. affinis using the mitochondrial genes 16S rRNA and cytochrome c oxidase 1 (CO1) revealed a significant genetic heterogeneity in the northern hemisphere [12,15]. These studies allowed one to define six main clades observed nearly all over the Holarctic, with striking levels of sequence divergence up to 10% in 16S rRNA and up to 19% in CO1. As a result, E. affinis was recognized as a cryptic species complex [15].
The phenomenon of cryptic speciation was supported by hybridization experiments that showed reproductive isolation among some North-American populations [12] and between North-American and European populations (S. Souissi, unpublished). Furthermore, significant eco-physiological differences between one North-American and one European population were found [16]. These studies provided the reason for detailed morphological analyses. Qualitative and quantitative differences of mandible, swimming legs, fifth rudimental leg and genital-double-somite structures were observed. Thus, E. cf. affinis from the Chesapeake Bay (USA) was described as a new species, E. carolleeae Alekseev and Souissi, 2011 and the Asian E. affinis from the Caspian Sea was also described as a new species, Eurytemora caspica Sukhikh and Alekseev, 2013.
Genetic structuring and high heterogeneity were also found among and within clades of the E. affinis species complex [6,12,20]. Genetic divergences within each clade were 1.7–12.4% in COI and 4–6% in 16S rRNA, and maximum pairwise divergences of 10% in 16S rRNA and 19% in COI were observed among clades. In Western Europe, three subclades were represented [6]: the NSEC (North Sea/English Channel) lineage, including populations from the Tamar, the Seine and Scheldt Estuaries, the Baltic lineage, including the specimens from Sweden and the East Atlantic lineage including specimens from the Loire and Gironde Estuaries. The population from the Gulf of Riga (the Baltic Sea) was composed of a mixture of haplotypes from both NSEC and East Atlantic lineages. Morphological comparison of populations from several European water bodies also revealed significant heterogeneity among populations [18].
In this study, we used a combination of morphological and genetic features to determine the intraspecific structure of the E. affinis complex in Western Europe. The specific objectives of the present paper were:
- • to compare the morphology of E. affinis among populations;
- • to determine whether morphometric results coincide with the genetic structure of the three European lineages found in populations throughout Western Europe.
2 Material and methods
2.1 Sampling
Animals were collected between 2005 and 2011 from 11 European sites: the river estuaries of Elbe, Seine, Scheldt, Tamar, Loire, Gironde, the Gulfs of Finland and of Riga, the Vistula Lagoon, the Swedish coast of the Baltic Sea and the Lake in the Bois-de-Boulogne (Paris) (Fig. 1). Samples were collected with plankton nets of 100- or 230-μm mesh-size from the surface layer (1–2 m deep) by boat or from shore and preserved in 96% ethanol. Some of these sites had already been studied by Winkler et al. [6] and were resampled again for morphological analysis and to evaluate the temporal stability of the genetic composition (the collection information for the specimens examined in this study is provided in Table 1).

Sampling locations of the Eurytemora affinis populations studied here. 1–3, Gulf of Finland (1, Gulf of Vyborg, 2, Neva Estuary, 3, Luga Estuary), 4, Gulf of Riga, 5, Vistula lagoon, 6, Baltic Proper (Sweden), 7, Elbe Estuary, 8, Scheldt Estuary, 9, Lake in the Bois-de-Boulogne (Paris), 10, Seine Estuary, 11, Tamar Estuary, 12, Loire Estuary, and 13, Gironde Estuary. The symbol ♦: corresponds to the sampling places studied in this work: ●: sampling places, the material which was used from previous work [6,12]; ■: sampling places, where the samples were taken twice for a previous study [6] and for the present study.
Mean values of selected morphometric indexes in females of the three lineages of E. affinis (Poppe, 1880) from their type localities.
| Lineage/Index | Caudal rami, L/W | Genital somite W1/W2 | P4 | P5 | Number of studied individuals |
| East Atlantic group | 6.9 ± 0.1 (5.1–8.9) | 1.2 ± 0.0 (1.1–1.3) | 1.2 ± 0.0 (1.1–1.3) | 2.7 ± 0.1 (2–4.4) | 34 |
| Baltic group | 8.2 ± 0.1 (6.4–12) | 1.1 ± 0.0 (0.9–1.4) | 0.9 ± 0.0 (0.8–1.1) | 2.8 ± 0.0 (2–5.1) | 99 |
| NSEC group | 6.1 ± 0.1 (5.0–7.8) | 1.3 ± 0.0 (1.1–1.5) | 1 ± 0.0 (0.8–1.1) | 2.4 ± 0.0 (1.8–3.4) | 37 |
2.2 Phylogenetic analyses
The sequence data used in this study were obtained from:
- • former studies [6,12] (GenBank accession numbers: JF727310–JF727558);
- • newly sequenced 198 individuals from six resampled populations and five new populations (see Fig. 1 and Supporting data, Table 1).
For a detailed description of sampling in 2006 and laboratory procedures, see Winkler et al. [6]. Newly sampled E. affinis were analysed as follows: the genomic DNAs were extracted from single adult copepods preserved in 96% ethanol using a standard method described by Aljanabi and Martinez [21] or using a cell lysis buffer with Proteinase-K protocol modified from Hoelzel and Green [17,22]. Universal primers COIH 2198 (5′-TAAACTTCAGGGTGACCAAAAAATCA-3′), COIL 1490 (5′-GGTCAACAAATCATAAAGATATTGG-3′; [23]) and specific EuF1 (5′-CGTATGGAGTTGGGACAAGC-3′), EuR2 (5′-CAAAATAAGTGTTGGTATAAAATTGGA-3′; [6]) were used for polymerase chain reaction (PCR) of COI amplification. Two types of temperature profiles modified after Lee [12] were used for PCR amplification. The first was: 5 cycles of 90 °C (30 s), 45 °C (60 s), 72 °C (90 s), followed by 27 cycles of 90 °C (30 s), 55 °C (45 s), 72 °C (60 s) ending with 5 min at 72 °C, and the second one starting with denaturation at 95 °C for 30 s, followed by 5 cycles of 90 °C (30 s), 55 °C (60 s), 72 °C (90 s), followed by 27 cycles of 90 °C (30 s), 55 °C (45 s), 72 °C (60 s) ending with 5 min at 72 °C. The product was purified with a Qiaquick PCR purification kit (Qiagen, Valencia, CA, USA), then, sequenced using an Applied Biosystems Inc. 3130 or 3100 automated sequencer (Applied Biosystems Inc., Foster City, CA, USA). Both DNA strands were sequenced to confirm the accuracy of each haplotype sequence.
The sequences were aligned using the algorithm CLUSTAL W [24] implemented in BIOEDIT v.7.0.3 [25] with manual editing of ambiguous sites. The obtained sequences were deposited in the GenBank database (accession numbers HM368364, HM473958–HM474035). Phylogenetic analyses were done with both the complete dataset of 447 sequences (198 original and 249 previously published; [6]) and with a reduced dataset containing only the 27 most frequent haplotypes (observed three or more times). Four sequences of E. carolleeae were sequenced in a previous study [8] and four sequences of Eurytemora lacustris were sequenced during this study and were used as outgroups.
Phylogenetic reconstructions were performed by Maximum Likelihood (ML) with PHYML v3.0 [26] and by Bayesian analyses with MrBayes 3.1 [27]. The best fitting model under the ML criterion was selected from the “Bayesian Information Criterion” (BIC) and “Akaike Information Criterion” (AIC) output of jMODELTEST v0.1.1 [28]. The ML analyses were conducted using a TPM1uf model with gamma rate distribution (G) and without invariable sites (I), the best tree topology was searched using NNI and SPR methods and node stability was estimated by 100 nonparametric bootstrap replicates. Two independent runs of 10 million generations each were performed for a Bayesian phylogenetic sampling every 1000th generation. The analyses were conducted using a TPM1uf model with gamma rate distribution (G).
The level of genetic differentiation in CO1 based using a Tamura–Nei 93 model was estimated in Mega 6 [29].
2.3 Haplotype diversity and network
Genetic diversity was compared among populations using DNASP v5.1 [30] including haplotype diversity (Hd, probability that two randomly chosen haplotypes are different in the sample; [31]) and nucleotide diversity (p, average number of nucleotide differences per location between two sequences; [32]). A median-joining haplotype network was constructed with the Network 4.6 program [32–34]. As two primer sets (EuF1–EuR2 and COIH 2198–COIL 1490) were used for the sequencing of different length fragments of the COI gene, a consistent fragment of 531 bp was chosen for the haplotype network analysis.
2.4 Morphological study
A total of 170 specimens were studied using morphological criteria. From 19 to 37 adults from each population were selected for morphological analyses. Before dissection, adult copepods were measured with an ocular micrometer (5 μm resolution). The dissection was processed in glycerol. After dissection, the specimens were placed on slides in pure glycerol, covered with a cover slip and ringed with Canada balsam.
The slides were then examined at maximum resolution up to 1000× (plan objective 100×, oil immersion) under a compound microscope (Zeiss IMAGER) equipped with a Nomarski system for differential interference contrast microscopy and a drawing tube. All measurements are given in μm.
Both sexes were studied for the Elbe estuary population, whereas only females were studied in all other populations. In total, 16 different characters that are commonly used in copepod taxonomy were measured. However, only eight of them were selected, which were most indicative statistically and correlated well with genetic lineages (Fig. 2).

Scheme, showing 8 characters measured in Eurytemora affinis (Рорре) female on (А) the caudal rami: length (L), width (W); the genital somite: W1, W2; (B) the distal segment of P4 exopodite: length of segment (LSeg) and distal spine (LongSp); (C) P5: length of distal spine (LongSp), spine 1 (Sp1).
Secondary sexual dimorphic characters typically used in copepod taxonomy were analyzed: the shape of the genital somite in females and the length of spines in the fifth legs. We measured the width of the genital-double-somite of the anterior (W1) and the posterior (W2) parts (Fig. 2A) and the spine lengths (LongSp, Sp1, Sp2, Sp3, TSp) as well as the distal segment length (Lseg) for the leg 5 exopodite (Fig. 2C). Additionally, the following features were selected: the caudal rami length (L) and width (W) (Fig. 2A); the distal exopodite segment (LSeg) as well as the distal spine length (LongSp) in the swimming leg 4 (Fig. 2B). The most taxonomically important indexes were chosen for further statistical analyses: caudal rami index (L/W), genital somite index (W1/W2), P4 LongSp/Lseg index, P5 LongSp/Sp1 index (Table 1). The characters used for index calculation are shown below in the “Results” section. A nonparametric Kruskal–Wallis test was performed with the standard statistical software Statistica 7. The significance level was set as P < 0.05.
The neotype slides for E. affinis were placed in the type collection of the Zoological Institute of the Russian Academy of Sciences, Saint Petersburg, Russia (ZIN RAS) under reference numbers 55080–55086. Copepod body and appendage terminology mainly followed Huys and Boxshall [35] and Alekseev and Defaye [36].
3 Results
3.1 Genetic study
Analyses of 436 CO1 gene sequences confirmed the presence of three lineages within the species complex of E. affinis in Europe [6]. Sequences from the Baltic Sea were mainly grouped in one lineage including sequences from the populations at the Swedish coast, in the Gulf of Finland, in the Gulf of Riga, in the Vistula lagoon and one sequence from the center of the Baltic Sea proper. The North Sea/English Channel lineage consisted of sequences from populations in the Elbe, Scheldt, Seine, and Tamar estuaries. The third lineage, the East Atlantic was mainly found in two locations: the Loire Estuary and the Gironde Estuary. Pairwise divergences consisted of 1.7% between Baltic and NSEC populations, 1.9% between Baltic and East Atlantic populations and 2.4% between NSEC and East Atlantic populations. The mean sequence divergence within each lineage was 0.1%, 0.3% and 0.5% in the NSEC, Baltic and East Atlantic populations, respectively. The three lineages were separated by on average 9, 10 and 13 mutational steps (mean group differences) for NSEC–Baltic, Baltic–East Atlantic and NSEC–East Atlantic, respectively (Fig. 3).
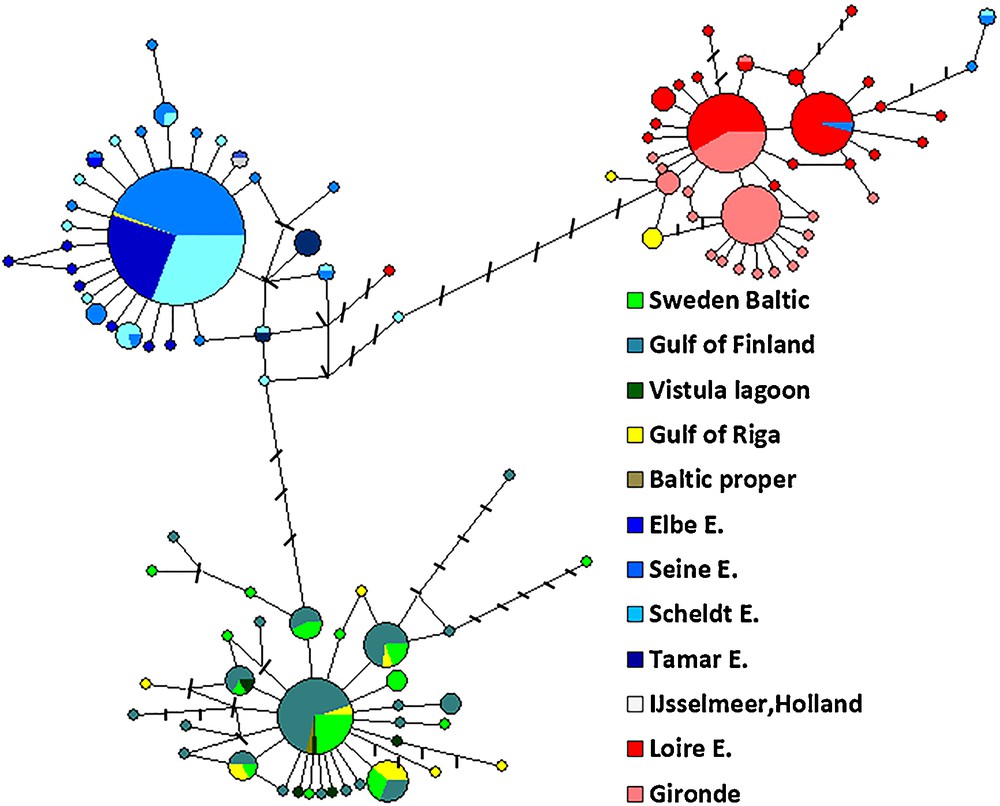
Parsimony median-joining network using cytochrome oxidase I (COI, 652 base pairs) showing the phylogenetic relationships among haplotypes in the European Eurytemora affinis. The size of the circles is proportional to the haplotype frequency. The smallest and the biggest circles represent 1 and 127 individuals, respectively. The numbers of mutations (greater than one) between haplotypes are indicated on the branches by black bars, which represent missing haplotypes.
Both haplotype (Hd) and nucleotide (p) diversities were higher in the Baltic lineage 0.874 ± 0.022 and 0.0045 ± 0.0025, respectively, than the NSEC lineage, which showed two times less haplotype and nucleotide diversities 0.481 ± 0.048 and 0.0022 ± 0.0018 and the East Atlantic lineage 0.829 ± 0.021 and 0.0031 ± 0.0021, respectively (Table 2).
CO1 haplotype variation of E. affinis populations at each location.
| Region | Sampling locations | N | Number of haplotypes | Hd ± SD | p(π) ± SD |
| Baltic | Gulf of Finland | 67 | 20 | 0.829 ± 0.038 | 0.0030 ± 0.0019 |
| Gulf of Riga | 14 | 8 | 0.868 ± 0.076 | 0.0044 ± 0.0025 | |
| Gulf of Riga other lineages | 5 | 3 | 0.700 ± 0.218 | 0.0984 ± 0.0049 | |
| Vistula Lagoon | 5 | 5 | 1.000 ± 0.126 | 0.0030 ± 0.0036 | |
| Sweden | 31 | 14 | 0.890 ± 0.040 | 0.0039 ± 0.0021 | |
| Baltic Sea, Proper | 1 | 1 | |||
| Overall within Baltic lineage | 123 | 38 | 0.874 ± 0.022 | 0.0045 ± 0.0025 | |
| North Sea/English Channel | Elbe | 40 | 10 | 0.404 ± 0.099 | 0.0010 ± 0.0017 |
| Scheldt | 54 | 13 | 0.478 ± 0.084 | 0.0022 ± 0.0013 | |
| Seine | 75 | 16 | 0.443 ± 0.073 | 0.0025 ± 0.0017 | |
| Tamar | 5 | 2 | 0.400 ± 0.237 | 0.0015 ± 0.0013 | |
| IJsselmeer | 1 | ||||
| Overall within NSEC lineage | 175 | 33 | 0.481 ± 0.048 | 0.0022 ± 0.0018 | |
| East Atlantic | Loire | 74 | 20 | 0.756 ± 0.036 | 0.0026 ± 0.0019 |
| Gironde | 62 | 18 | 0.757 ± 0.041 | 0.0024 ± 0.0018 | |
| Overall within East Atlantic lineage | 136 | 36 | 0.829 ± 0.021 | 0.0031 ± 0.0021 | |
| Overall | 436 | 105 | 0.888 ± 0.011 | 0.0143 ± 0.0026 |
Generally, lineages were well separated geographically. However, some mixing among lineages was noted. In the Gulf of Riga, five individuals (2006 year sampling) out of 19 belonged to the East Atlantic and NSEC lineages; the 14 individuals collected in the City Port in 2008 belonged to the Baltic lineage. Furthermore, in 2011, in the Seine Estuary, near Tancarville, two individuals out of 37 and in the Scheldt Estuary near Antwerp 1 individual belonged to the East Atlantic rather than to the NSEC lineage. In the same year 2011 in the Loire Estuary, one individual out of 52 belonged to the NSEC lineage (Fig. 3). In a previous work [6], only one haplotype out of 38 from the Seine Estuary was nonnative (East Atlantic) since its habitat and mixing was not revealed in the Loire Estuary.
We found 105 distinct haplotypes among the sequenced 436 individuals. Most of the haplotypes were private. The haplotype network for the NSEC lineage was star-like, centered around the dominant haplotype, whereas the haplotype network in the Baltic and East Atlantic lineages were less star-like and more dispersed (Fig. 3). The dominant haplotype in the Baltic lineage was mainly one mutational step away from the less frequent haplotypes.
Three closely related dominant haplotypes were observed in the East Atlantic lineage, the most frequent haplotype occurred in both estuaries, whereas the two less dominant haplotypes were only one mutational step away from the dominant and either present in the Loire or the Gironde estuary.
The Bayesian and the Maximum Likelihood trees (Fig. 4) presented similar topologies at the higher nodes with strong supports separating the three different lineages.

The phylogenetic tree constructed by Bayesian analysis using TPM1uf + G model on the 27 most frequent haplotypes using cytochrome oxidase I (COI, 652 base pairs). Bayesian posterior probabilities (BPP) and bootstrap values for Maximum Likelihood (ML) (≥ 60%) are given for nodes. Numerals, following abbreviations indicate the frequency of this particular haplotype in the samples. Eurytemora carolleae and Eurytemora lacustris are used as outgroups. Abbreviations: GF: Gulf of Finland; Sw: Sweden; GR: Gulf of Riga; BP: Baltic proper [12]; VL: Vistula lagoon; L: Loire Estuary; G: Gironde Estuary; P: lake in Bois-de-Boulogne, Paris; S: Seine Estuary; Sch: Scheldt Estuary; E: Elbe Estuary.
3.2 E. affinis (Poppe, 1880) morphological heterogeneity
The morphological variability among the populations was estimated in a number of sites: Gironde, Loire and Seine Estuaries, France; Elbe Estuary, Germany; Luga Estuary, Gulf of Finland and Vistula Lagoon, Baltic Sea, Russia; Gulf of Riga, Baltic Sea, Latvia. Similar to molecular genetic analysis, morphological variability was not random, but grouped in accordance with three main regions (NSEC, Baltic, East Atlantic lineages) (Figs. 5 and 6).
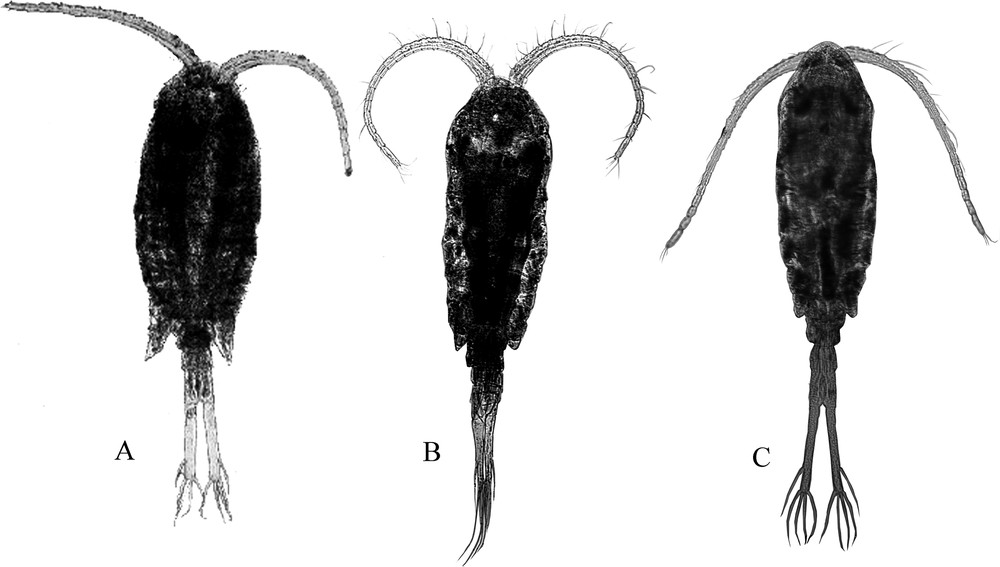
Eurytemora affinis (Poppe) female (A) from the Elbe Estuary, (B) from the Gironde and (C) from the Luga Estuary (Baltic Sea).
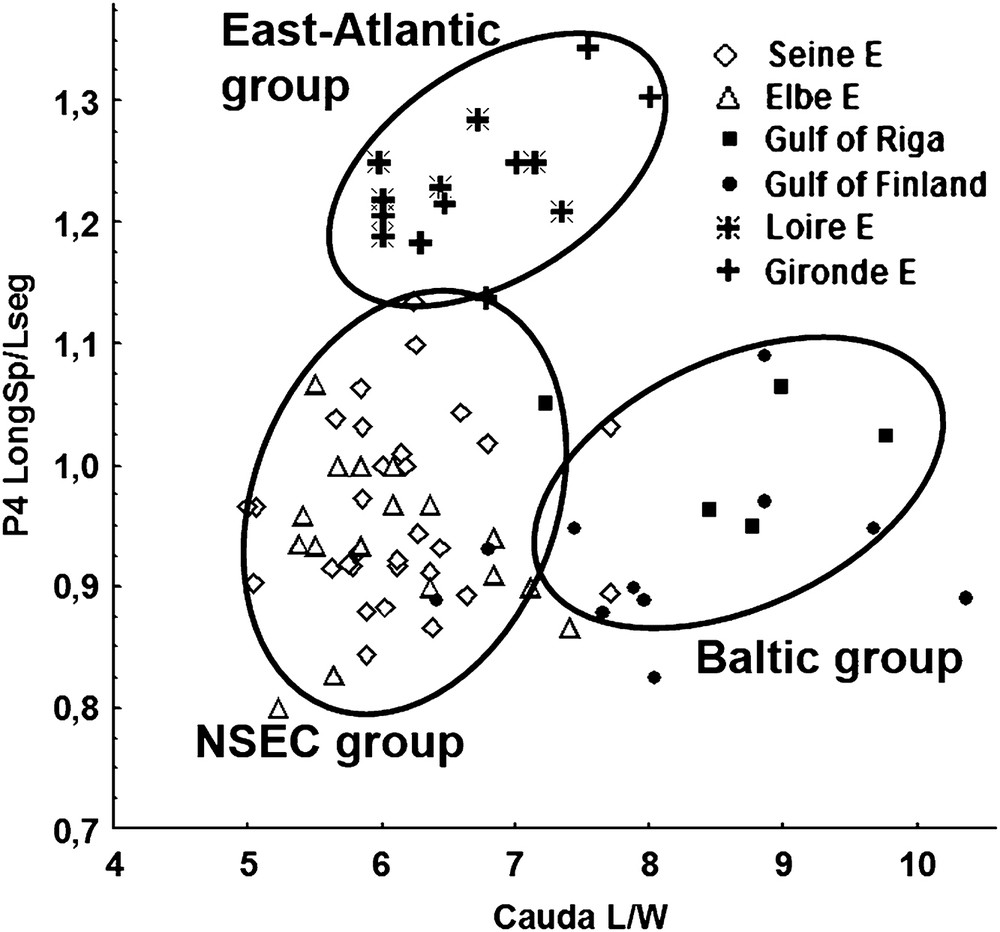
Morphological differences between the populations of Eurytemora affinis females from the East Atlantic (Loire, Gironde estuaries), the North Sea Basin (Seine, Elbe estuaries), and the Baltic Sea (Gulf of Finland, Gulf of Riga). Chart based on caudal index [ratio caudal length (L)/width (W)] and the ratio of the spine 1 length/distal segment length P4.
The differences among the groups of Е. affinis were based in the females on measurements of caudal rami, genital somite, and some structures in P4 and P5 (Table 1). Caudal index L/W distinguished consistently among all these groups, genital somite indices W1/W2 differed consistently among the Atlantic–Baltic and the NSEC–Baltic groups, the index P5 LongSp/Sp1 differed between the Atlantic–NSEC and the NSEC–Baltic groups, the index P4 LongSp/Lseg differed between the Atlantic–NSEC, and the Atlantic–Baltic groups.
Overall, the specimens from the Elbe Estuary (NSEC lineage) had a shorter distal spine at P5 and the genital somite had a constriction in the middle part, like E. carolleeae, but the wing-like outgrowths were rather small. Females from the Gironde Estuary (East Atlantic lineage) and from the Gulf of Finland (Baltic lineage) had a genital somite of more cylindrical shape. The individuals from the Elbe Estuary showed long and strong hair-like setae on both sides of the caudal rami, as well as on the last abdominal somite. The hair-like setae in E. affinis from the Baltic Sea were not so dense. The Baltic specimens generally had longer caudal rami: the mean ratio length/width was 8.2. in E. affinis from the Gironde with barely noticeable spines on the caudal rami and a long apical spine on the P4 exopodite distal segment. The specimens from the Loire and Gironde estuaries often had segment-like divisions of swimming leg setae, but this seta segmentation was not as strong as described for E. carolleeae [18] and was rather similar to that in E. caspica [7]. Possibly, the seta segmentation is a rather primitive character, which indicates a more ancient population of E. affinis, which is more closely related to the American Eurytemora. This suggestion is in accordance with the genetic data, which showed, that the Loire-Gironde population represents a more ancient group within the European populations.
The differences among populations within the East Atlantic, the Baltic and the NSEC lineages, were insignificant or less than between the groups.
The genetic analyses of 436 CO1 gene sequences and the morphological searches of 170 specimens revealed that only E. affinis s. str. is present at most of the studied places. Besides E. affinis, its sister species E. carolleeae was observed in the Gulf of Finland and in the Gulf of Riga. These findings were described earlier in Alekseev et al. [4] and Sukhikh et al. [8] and not discussed here.
4 Discussion
4.1 Genetic study
Genetic heterogeneity among E. affinis populations in Western Europe were analyzed in detail in a previous article [6]. Using a more comprehensive sampling scheme within the Baltic Sea by increasing the number of analyzed specimens confirmed previous results [6]. Moreover, our results more fully elucidated the patterns of differentiation among the European lineages of E. affinis and especially clarified better the genetic structure of populations within the Baltic lineage.
The haplotype network reflected the relative geographic position of the lineages. The NSEC lineage was situated in the center between the Baltic and the East Atlantic lineages. In general, all lineages had more or less star-like patterns centered around dominant haplotypes, and were associated with unimodal pairwise haplotype mismatch distributions (results not shown). Such results are compatible with a recent population bottleneck or the founder event as an effect of a more recent expansion [37–39], that possibly happened after the last deglaciation (18,000 years BP). For the NSEC lineage, this inference was supported also by low haplotype and nucleotide diversities. Despite the fact that in this work we studied almost two times more number of CO1 sequences, the above-mentioned results on the shape of haplotype networks and levels of haplotype and nucleotide diversities corresponded globally to the data discussed by Winkler et al. [6]. The only exception was that the haplotype network in the Baltic lineages were less star-like and more dispersed in the present work.
The phylogenetic analysis of the European E. affinis using CO1 confirmed the existence of three lineages (Baltic, NSEC and East Atlantic) with a clear geographical pattern in the lineage structure. There were sporadic exceptions, which could be the result of historical events and/or recent human activities – as all studied areas are major centers of maritime traffic. The three sampling stations in the Gulf of Riga (the Baltic Sea), an area with high shipping activity, revealed the presence of all three lineages. The city port station was inhabited by the Baltic lineage (14 specimens), whereas the NSEC (one specimen) and East Atlantic (four specimens) were observed in deeper off-shore stations, that were probably introduced in the Gulf of Riga with the ballast water of ships. According to international regulations, ballast water of ships should be exchanged in the open part of the sea [40], so that this might be the reason why we found nonnative NSEC and East Atlantic Eurytemora at the entrance to the Gulf of Riga. Co-occurrence of NSEC and East Atlantic haplotypes was also observed in the Loire, the Seine and the Scheldt estuaries.
Compared to a previous study (sampling 2006 from Winkler et al. [6]) there was an increase in the nonnative haplotypes observed in the Seine Estuary. Different co-occurring lineages were firstly revealed in the Loire and the Scheldt estuaries in the present study (2011 sampling year) compared to earlier records (2006 [6]). Possibly, this is a result of continuing Eurytemora invasions with ship ballast waters.
Interestingly, neither NSEC nor East Atlantic haplotypes were found in the eastern part of the Gulf of Finland. One of the biggest Baltic cargo side ports in Saint Petersburg is situated there. Many invasive species occupied this water area, and the North-American E. carolleeae was found here a few years ago [4,8], whereas the presence of mixed Eurytemora populations in the Loire, Seine, Scheldt, Riga estuaries resulted from recent human activities; a relatively low salinity 1.5–5‰ in the Gulf of Finland (compared to other studied areas) could possibly be the main reason for a single Baltic lineage present there only. Possibly, East Atlantic and NSEC populations prefer more saline conditions, close to native habitats. Probably the study of the Gulf of Finland marine part will reveal a mixture of lineages there. In turn, no Baltic specimen was observed in the East Atlantic and NSEC water areas. This could be also the result of different salinity preferences. At the same time rather stressful and unusual tidal conditions of the North Sea and the Bay of Biscay are, possibly, also a barrier for the Baltic Eurytemora.
Our population genetic study of E. affinis from the inner waters in the lake in the Bois-de-Boulogne (Paris) revealed the presence of the East Atlantic lineage. The lake is connected to the Seine River, thus, we expected a dominance of the NSEC lineage, which prevailed in the Seine Estuary. However, the number of sequenced specimens was very low (n = 3). More data will be needed to test whether central France might be a sympatric area or a geographic border for NSEC and East Atlantic populations. A similar picture was observed in E. carolleeae in the Pacific Northwest, the Northeast Atlantic along the North-American continent and the phylogeographic break in Britany, France [6,12,20], or if the occurrence of specimens of the East Atlantic lineage might be the result of human activity.
4.2 Morphological characters
E. affinis (Poppe, 1880) is a broadly distributed species with huge morphological variability. Multiple species and subspecies were described within the boundaries of this species almost from the moment of its first description: E. affinis hispida (Nordquist, 1888), E. affinis hirundo Giesbrecht, 1881, E. raboti Richard, 1897, E. hirundoides (Nordquist, 1888). Nowadays only E. raboti is valid. All other species are not recognized by most researchers of the scientific community, as it is difficult to distinguish what type of variability was described in these taxa–genetic or just phenotypic plasticity. It is interesting to note that most of them were distinguished within European waters. The performed morphological observation of European populations from the Gironde, Loire and Seine Estuaries, France from the Guadalquivir River in Seville, Spain, from Helsinki's Fishing Port, Finland; from the Luga Estuary, Gulf of Finland, Baltic Sea, Russia, from the Gulf of Riga, Baltic Sea, Latvia, from the Scheldt Estuary, Belgium, from the Elbe River, Germany, from the Seine and Loire Rivers also showed a substantial variability between these populations [18,41].
Our genetic studies of these populations revealed three lineages, corresponding to three geographical regions: East Atlantic, Baltic and North Sea–English Channel. From 1.4 to 2.4% of nucleotide substitutions were observed among them. Such genetic differences among European lineages of E. affinis occurred at the subspecies level. As the mean sequence divergences within species in the genus Eurytemora were 19–25% in the CO1 region and 10% in the 16SrRNA region [12], while the mean genetic differences in the mitochondrial gene 16SrRNA among species of the genus Calanus were 12% to 18% and subspecies differences between Calanus pacificus oceanicus and C. p. californicus were a 0.9%–1.0% sequence divergence [42]. Fine morphological analyses of copepods from the Elbe, Seine, Gironde, Loire estuaries and from the Baltic Sea revealed a picture fully consistent with the genetic results. Significant morphological differences were observed among the populations in measures of the genital somite, the fourth pairs of legs (P4), the caudal rami. Such differences were congruent with genetic differences among populations and allowed one to identify three morphological groups within the European E. affinis. The levels of genetic and morphological heterogeneity among these groups corresponded to the subspecies level. However, for subspecies description, one needs to estimate the stability of the morphological differences in different populations and for both sexes according to the regulations of the Zoological Nomenclature.
Morphological variability (error/mean) within studied groups was rather low and never exceeding 3.6%. The maximal variability in most indexes (Caudal rami L/W, Genital somite W1/W2, P5 LongSp/Sp1) was observed within the Loire–Gironde populations. At the same time, the Baltic populations showed mean morphological variability (Caudal rami L/W, Genital somite W1/W2, P4 LongSp/Lseg), the values of which were minimal for the North Sea group. This completely corresponds to levels of genetic variability among CO1 clades. Moreover, it does not contradict the assumption, described in Winkler et al. [6], that the East Atlantic lineage was separated earlier and may have survived in the proposed southern glacial refuge without strong selection. NSEC lineages may have been preserved in some northern glacial refugia and went through population bottlenecks. High genetic, morphological heterogeneity and dispersed haplotype networks of the Baltic group have been probably caused by surviving recent glaciations separated from the two other lineages in a few different refuges. The genetic data showing that the East Atlantic population is more ancient among all European populations is also supported by primitive features such as seta segmentation, which was described in the American sister species E. carolleeae and in the Caspian species E. caspica [7,18].
Attempts at obtaining morphological data with the existing descriptions of different E. affinis-group species could provide interesting results. However, there were no type specimens for comparison with the present material. Moreover, the only type exemplar of E. affinis species from the Elbe River was lost. Nonetheless, given pictures and description of the type exemplar of E. affinis (Poppe, 1880) fully corresponds to our studied E. affinis from the Elbe River.
We can also suppose that according to the pictures and places of study, E. hirundoides described by Nordquist in 1888 [43] from Finnish waters most likely corresponds to E. affinis from the Gulf of Finland. E. hirundo, described by Giesbrecht in 1881 [44] from Kiel and observed in the Kattegat, looked like an intermediate form between E. affinis from the Elbe estuary and E. affinis from the Baltic Sea. Considering the geographic position of this species, it could actually represent a mixed population between relatively isolated forms from the Baltic and North Sea.
We did not obtain material from the Kiel bay to analyze the populations there, but every studied lineage looked more or less homogeneous, without intermediate forms. As the mitochondrial-DNA analysis did not show whether there were hybrids within the sampled population, a study of nuclear genes and crossbreeding experiments could clarify this issue.
As for East Atlantic populations, a number of authors studied the Gironde population and identified Eurytemora specimens as E. affinis hirundoides in their publications [45,46]. Possibly, it was because of a more slender common view comparing with E. affinis from the type locality. More precise analyses of this form were impossible, as the figures in the original description were of rather low resolution.
In this regard and as genetic and morphological data were in accordance with each other and present three restricted groups of organisms, three local populations within the European E. affinis were set: E. affinis s. str. from the Elbe estuary, E. affinis from the Gironde River, and E. affinis from the Baltic Sea.
5 Conclusions
(1) Three local populations observed in this work possibly appeared as a result of some historical events and of geographic isolation. Shared haplotypes among the studied Baltic, NSEC and East Atlantic lineages possibly reflects a high level of ship traffic and ballast water exchange during the 20th century.
(2) Both morphological and genetic searches were basically congruent and revealed that the East Atlantic population was separated from other populations first.
(3) The morphological and genetic differences observed among these three local E. affinis populations as well as their geographically isolated areas correspond to subspecies level in the sense of the Code of Zoological Nomenclature. Some additional studies on morphologically stable differences among these populations are planned in the near future to describe them as new taxonomical units.
Acknowledgements
We are very grateful to M. Tackx and O. Glippa for their help in sampling the Elbe, the Scheldt, the Seine, the Gironde and the Loire estuaries. We thank C.E. Lee for providing E. affinis sequences for comparison and discussion purposes. We thank the whole staffs of the GEPV laboratory, Lille-1 University, and of the Laboratory of Molecular Genetic Systematics, Zoological Institute of the Russian Academy of Sciences, Saint Petersburg, where the genetic part of the work was done, and the Canadian Center for DNA Barcoding for the partial material sequencing, which was funded by Genome Canada, through the Ontario Genomics Institute. For this work, the Federal Collection No. 96-03-16, Zoological Institute of the Russian Academy of Sciences, Saint Petersburg, was used. This work was supported by the BIODISEINE and ZOOGLOBAL Seine Aval projects in France, the “Biodiversity” grant from the Presidium of the Russian Academy of Sciences; Russian Foundation for Basic research under grants Nos. 14-04-01149 А, 14-04-00932 A and 16-34-00086 мол_а. We acknowledge critical comments by Hans-Uwe Dahms that improved an earlier version of this manuscript.


