1 Introduction
Bacterial blight (BB), caused by Xanthomonas oryzae pv. oryzae, is a most destructive disease of rice reported from Asia, northern Australia, Africa, and the Caribbeans. In Asia, including Bangladesh, millions of hectares of rice fields are severely affected every year, with reported yield losses of up to 60 per cent. The bacterium causes yellowing and drying of leaves and wilting at the seedling stage, while blight lesions caused by severe strains giving a striped appearance on the leaves and field patches infested with BB have a whitish, ragged appearance. Some cultural, chemical and biological control approaches have been developed to control the disease, but none of them has been found to be fully effective in case of outbreaks. Therefore, the use of resistant cultivars is the most promising method for the control of the disease. Most of the resistant genes provide an effective and stable protection against subpopulations of the pathogen. But changes in the pathotype structure within the population break down the resistance, which may result from several factors such as genetic change or migration from other geographical areas.
A high degree of genetic diversity and pathotypic variation of X. oryzae pv. oryzae strains was reported from major rice-growing countries of Asia such as China, India, Indonesia, Korea, Malaysia, Nepal, Sri Lanka, and the Philippines [1,3,18]. There are over 30 reported races from several countries [1,13,17]. A repetitive DNA element cloned from X. oryzae pv. oryzae [10], several transposable elements and an avirulence gene were used to assess the population structure and the genetic diversity of strains of X. oryzae pv. oryzae collected between 1972 and 1988 from the Philippine Islands [16]. To expedite the analysis of a large number of samples, a simple and efficient DNA fingerprinting method based on polymerase chain reaction (PCR) using primers corresponding to the repetitive element IS1112 was developed [7,19]. A set of races identified in the Philippines using five differential rice cultivars has been used widely for identifying and classifying resistance to BB in other cultivars [12,13]. Most strains from South Asia (Nepal and India) were virulent to cultivars containing the BB resistance gene Xa5, while most strains from other countries were avirulent to Xa5. The regional differentiation of clusters of X. oryzae pv. oryzae in Asia and the association of some pathotypes of X. oryzae pv. oryzae with single clusters suggested that strategies that target regional resistance breeding and gene deployment are feasible [3]. The pathogenic diversity of X. oryzae pv. oryzae strains from the Yunnan Province of China revealed that the strains were polymorphic for virulence to 12 near-isogenic lines [17]. The assessment of the genetic and pathogenic diversity of X. oryzae pv. oryzae on a high-yielding local variety in India revealed that all of them were compatible with the resistance genes, while these pathotypes were incompatible with the genes, Xa5, Xa10, Xa13 and Xa21, indicating the possibility of deploying them for enhancing the resistance [19]. Recently, several pathotypes of X. oryzae pv. oryzae were identified based on the pathogenic variation on NILs of rice from the major rice-growing areas in Bangladesh [9]. Intriguingly, no comprehensive report on the field-based assessment of rice BB and of the genetic diversity of the bacterium, X. oryzae pv. Oryzae, has been reported yet in Bangladesh. In this study, 19 rice-growing districts were surveyed to know the status of BB disease during the rain-fed and irrigated seasons. The pathotypic and genetic diversity of the BB pathogen, X. oryzae pv. Oryzae, were studied to unravel the complex relationship between pathotypes and genotypes of the BB pathogen, X. oryzae pv. oryzae.
2 Materials and methods
2.1 Collection of BB infected rice leaf samples
Bacterial-blight-infected plant samples showing typical symptom were collected randomly from 19 different districts of Bangladesh (Fig. 1). Five to ten different locations of each district were selected for sampling. From each location, three upper leaves of 20 plants were collected, which formed a composite sample, and a representative sample was taken for the isolation of X. oryzae pv. oryzae. After collection, the diseased plant samples were brought in the laboratory and preserved in the refrigerator for isolation. A total of 96 isolates (from 127 samples) were purified.
Pathotypic distribution of X. oryzae pv. oryzae in 19 districts of Bangladesh.
2.2 Isolation of X. oryzae pv. oryzae isolates
Each diseased leaf sample was cut into small pieces, about 1–2 cm in length with the margin of typical lesions. The cut leaf pieces were sterilized in a 10% Clorox solution for 1 min and then with 70% ethanol for 1 min, followed by rinsing with sterilized deionized water. Each leaf sample was then homogenized with 1 ml of sterile distilled water. The resulting suspension was diluted serially (10−5 to 10−6) and then spread on a Nutrient Broth Yeast Extract (NBY) agar medium. The plates were then be incubated at 28 °C for 3–4 days.
2.3 Identification and purification of X. oryzae pv. oryzae cultures
The bacterial growth obtained in the plates was recorded and those having the morphological characteristics of Xanthomonas oryzae pv. oryzae were picked up with a sterilized loop, and purified cultures were obtained by streaking on both NBY and YDC (Yeast Dextrose Carbonate) agar media.
2.4 Confirmation of X. oryzae pv. oryzae isolates by the pathogenicity test
The isolates of X. oryzae pv. oryzae were confirmed by the pathogenicity test using two susceptible rice cultivars, TN1 and IR24. Seeds of the rice cultivars were sown in the plastic pot for raising the seedlings. The 20–25-day-old seedlings were then be transplanted into an earthen pot in the net house for inoculation. The bacterial isolates were cultured in NBY agar medium at 28 °C for 48–72 h and then resuspended in sterile distilled water at a cell density of 1 × 108 cells/ml measured by spectrophotometery. The second or third youngest leaf of each plant was inoculated using the scissors clipping method. Briefly, the tips of the leaves were clipped by a sterilized scissor dipped in the bacterial cell suspension. After inoculation with each isolate, the inoculated plants were kept in the net house for the development of symptoms.
2.5 Determination of pathotypes of Xanthomonas oryzae pv. oryzae isolates
Pathotypic analyses of X. oryzae pv. oryzae with a set of near-isogenic lines of rice viz. IRBB1, IRBB2, IRBB3, IRBB4, IRBB5, IRBB7, IRBB8, IRBB10, IRBB11, IRBB13, IRBB14 and IRBB21 carrying a single known BB resistance gene in the background of the susceptible cultivar IR24 were carried out. A susceptible check cultivar IR24 was maintained. Plants were raised in earthen pots; there were three hills in each pot. Three pots were maintained as replications for each isolate. Around 50-day-old plants were inoculated with the X. oryzae pv. oryzae isolates strain following the scissors clipping inoculation method [8]. The pairs of scissors was dipped into the bacterial suspension and the tips of the leaves were clipped. An individual isolate was used for inoculation on all the leaves of five plants which amounted to a total of around 20 leaves. The plants were cultivated in a net house under natural photoperiodic conditions. Twenty-one days after inoculation, the lesion length formed due to inoculation was measured with a ruler. The different X. oryzae pv. oryzae isolates were classified into pathotypes/races based on their virulence on the differential NILs. The samples showing a lesion length < 3 cm were considered as resistant and those showing a lesion length > 3 cm were considered as susceptible [3].
Assessment of genetic diversity ofX. oryzaepv.oryzaeisolates. Outwardly directed oligonucleotides complementary from each end of the IS1112 (a repetitive element isolated from X. oryzae pv. oryzae) was used to fingerprint DNA from the isolates of X. oryzae pv. oryzae, as described previously by George et al. [6] and Reddy et al. [19].Genomic DNA isolation. Genomic DNA of 96 X. oryzae pv. oryzae isolates was extracted from 1.5 ml of NBY or PS broth cultures grown overnight using the Gen Elute TM Bacterial Genomic DNA kit (Sigma Aldrich). The DNA samples were dissolved in sterile distilled water or a TE buffer (pH 8.0) and were preserved at −20 °C for immediate use. Five microliters of DNA extracted from each sample of the X. oryzae pv. oryzae isolates was run on 0.7% agarose gel along with 1 μl of DNA marker. The DNA concentration was then be estimated by comparing the glowing intensity of the sample DNA bands with the marker bands. For quantification of DNA concentration, the spectrophotometer wavelength was set at 260 nm after the spectrophotometer UV lamp was warmed up. A square cuvette (the zero or blank cuvette) was filled with 2 ml of double distilled water and placed in the cuvette chamber, then absorbance reading was adjusted to zero for standardization. The test samples were prepared by taking 2 μl of each DNA sample in the cuvette containing 2 ml of sterile distilled water, then mixed comprehensively by pipetting, placed in a spectrophotometer; the absorbance was read at 260 nm. Then the cuvette was rinsed out with sterile water, stamped out on a paper wipe, and the absorbance was recorded for each sample in the same way. Using the above absorbance measurements, the original concentrations were determined according to the following formula:
Construction of dendograms and phenograms. The banding patterns of each isolate was recorded in binary form with a number, 1 representing the presence and 0 representing the absence of each band, as analysed by the alphaimage software. The dendograms were constructed within the SAHN program of NTSYS-pc (Exeter software, Setauket, NY) using the unweighed pair–group method using arithmetic averages (UPGMA). The phenogram of X. oryzae pv. oryzae strains was based on the virulence to 12 near-isogenic lines containing a single gene for resistance. A data matrix was generated for the virulence data by scoring avirulence as 0 and virulence as 1. From these data, a phenogram was reproduced by the unweighted pair–group method for arithmetic average in the SHAN program (NTSYS-pc, version 1.80, Exeter Biological Software, Setauket, NY). Bootstrap analyses were performed using the computer program Winboot to assess the robustness of the dendogram.
3 Results
Determination of pathotypes and their reactions against NILs. All 96 Xoo isolates induced the typical disease symptom on susceptible check varieties IR24 and TN1. Pathotypic analyses of all Xoo isolates were conducted based on their reactions against 12 NILs and a total of 24 pathotypes/races were recorded. All isolates formed yellow mucous colonies on both NBY and YDC media. Pathotypes VI, XII and XIV consisted of maximum isolates (9) followed by pathotype XIII (8), pathotypes II and XVI (7), pathotypes I, IV and XI (4), pathotypes V, XV, XVII, XX, XXI and XXII (3 isolates each), pathotypes III, VI, VIII, IX, X, XVIII, IX and XXIII (2 isolates each). Pathotype XXIV is represented by a single isolate (Table 1).
Pathotypes of X. oryzae pv. oryzae in Bangladesh.
| Pathotypes | Near isogenic lines (NILs) with known resistance gene | |||||||||||||
| No. of isolates | % of isolates |
IRBB1 (Xa1) |
IRBB2 (Xa2) |
IRBB3 (Xa3) |
IRBB4 (Xa4) |
IRBB5 (Xa5) |
IRBB7 (Xa7) |
IRBB8 (Xa8) |
IRBB10(Xa10) | IRBB11(Xa11) | IRBB13(Xa13) | IRBB14(Xa14) | IRBB21(Xa21) | |
| I | 4 | 4.17 | S | S | S | S | S | S | S | S | S | S | S | S |
| II | 7 | 7.29 | S | S | S | S | S | S | S | R | R | R | S | S |
| III | 2 | 2.08 | S | S | S | S | S | S | S | S | R | R | S | R |
| IV | 4 | 4.17 | R | R | R | S | S | S | S | S | S | S | S | S |
| V | 3 | 3.13 | S | R | S | S | R | R | S | S | S | S | S | S |
| VI | 2 | 2.08 | S | R | S | S | S | S | S | S | S | R | R | R |
| VII | 9 | 9.38 | R | R | S | S | R | S | S | S | S | S | R | R |
| VIII | 2 | 2.08 | S | S | S | S | S | S | S | R | R | R | R | R |
| IX | 2 | 2.08 | R | S | R | R | R | S | S | R | S | S | S | R |
| X | 2 | 2.08 | R | R | R | R | R | R | S | S | S | S | S | R |
| XI | 4 | 4.17 | S | S | S | R | R | R | S | R | R | S | R | R |
| XII | 9 | 9.38 | S | S | R | S | R | R | S | R | R | S | R | R |
| XIII | 8 | 8.33 | R | R | R | S | S | R | S | R | S | R | R | S |
| XIV | 9 | 9.38 | R | R | S | S | R | S | S | R | R | R | R | R |
| XV | 3 | 3.13 | R | R | R | R | R | R | S | S | R | R | S | S |
| XVI | 7 | 7.29 | R | R | R | R | R | R | S | R | R | R | R | R |
| XVII | 3 | 3.13 | S | R | S | R | R | R | R | R | R | R | S | R |
| XVIII | 2 | 2.08 | S | S | R | R | R | R | R | S | S | R | S | S |
| XIX | 2 | 2.08 | R | R | R | S | R | S | R | R | R | R | R | R |
| XX | 3 | 3.13 | S | R | R | S | R | S | R | S | S | R | R | R |
| XXI | 3 | 3.13 | S | R | R | R | R | R | R | R | R | R | S | R |
| XXII | 3 | 3.13 | R | R | R | S | S | R | R | S | R | R | R | R |
| XXIII | 2 | 2.08 | R | R | R | R | R | R | S | R | R | S | R | S |
| XXIV | 1 | 1.04 | R | S | R | R | R | S | R | R | R | R | R | R |
All 24 pathotypes showed distinct virulence and avirulence reactions from each other with the NILs tested. The frequent distributions of a maximum of 14 pathotypes were recorded from the greater Mymensingh district, whereas the lowest single pathotype was recorded from the Natore and Gaibandha districts.
3.1 DNA finger printing analyses of Xanthomonas oryzae pv. oryzae isolates
To dissect the genetic variation among the isolates of X. oryzae pv. oryzae obtained from some selected rice-growing districts of Bangladesh, a subpopulation of 96 single mother colony isolates were obtained from high-yielding modern rice varieties, Swarna and hybrid rice varieties grown in farmer field during the irrigated and rain-fed seasons of 2014. The analysis of DNA fingerprints of these isolates was revealed separately by constructing the dendrogram. The DNA fingerprinting of 96 isolates of X. oryzae pv. oryzae was done using the PCR-based IS1112 marker with specific primers JEL-1 and JEL-2, which produced multiple DNA amplification products in the range of 0.2 to about 7 kb (Fig. 2 A, B, C, D, E & F), although the bands were lying close together due to the short running time of the gels. A total of 6–30 bands were amplified from the isolates of X. oryzae pv. oryzae. However, the number of distinguished scorable bands ranged from 7 to 18, out of which 10 bands ranging from 0.4 to 7 kb were found polymorphic among the isolates that were found out by the analyses with Alphaimage software. The other bands were common in all the pathotypes studied.
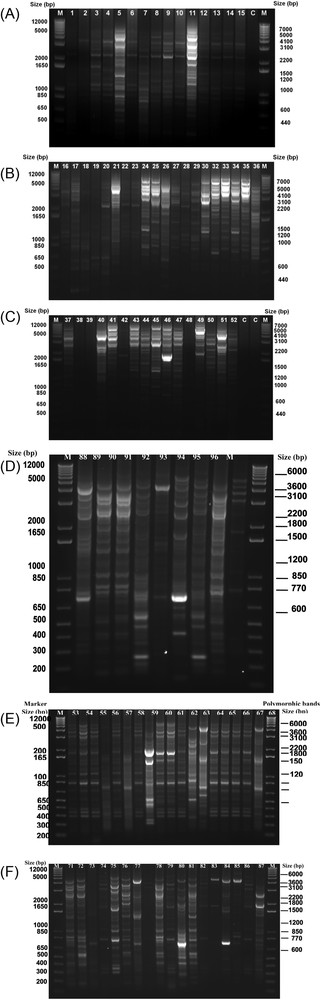
DNA fingerprint patterns of X. oryzae pv. oryzae isolates generated with IS1112-based primers (JEL1/JEL2) by PCR. Lane M represents the 1 Kb plus ladder, followed by A to F Lanes, 1–96 containing the fingerprints of X. oryzae pv. oryzae isolates (BXO1-BXO96) and C-control, collected from 19 rice-growing districts of Bangladesh during the irrigated and rain-fed seasons of 2014. The band sizes in the left-hand sight of each gel indicate the 1 kb plus ladder, and the band sizes in the right sight of each gel indicate the polymorphic bands identified by Alphaimage software among the X. oryzae pv. oryzae isolates. Masquer
DNA fingerprint patterns of X. oryzae pv. oryzae isolates generated with IS1112-based primers (JEL1/JEL2) by PCR. Lane M represents the 1 Kb plus ladder, followed by A to F Lanes, 1–96 containing the fingerprints of X. oryzae pv. ... Lire la suite
The dendrogram was constructed with UPGMA using the data sets obtained with the PCR-DNA fingerprinting method using the primers specific to the IS1112 elements presented in Fig. 3. Grouping based on the IS1112 primer amplifications revealed that all 96 Xoo isolates were distributed into 17 clusters along with 17 molecular haplotypes at around 65% level of similarity index. Among these clusters or haplotypes, I considered as major consisted of 46 isolates, followed by VII (7); III & IV (5); XIII (4); II, V, XIV & XV (3); VI, VIII, IX, & XVI (2), whereas X, XI, XII & XVII represented by single isolate.

Dendrogram constructed with UPGMA on the basis of the IS1112 repetitive element based on the polymerase chain reaction DNA fingerprint data analyzed with NTSYS software for a collection of 96 isolates of X. oryzae pv. oryzae from farmer field of 19 rice-growing districts of Bangladesh. The similarity percent is indicated by a coefficient. The dotted line indicates the similarity index level for clustering among the X. oryzae pv. oryzae isolates. At 65% similarity levels, I–XVII clusters and H1–H17 Molecular haplotypes were detected. Masquer
Dendrogram constructed with UPGMA on the basis of the IS1112 repetitive element based on the polymerase chain reaction DNA fingerprint data analyzed with NTSYS software for a collection of 96 isolates of X. oryzae pv. oryzae from farmer ... Lire la suite
Relationship between pathotypes and the haplotypes. A phenogram was constructed based on the virulence data of X. oryzae pv. oryzae isolates to the 12 near-isogenic lines of rice containing a single gene for resistance to interpret the relationship between the pathotypes and the molecular haplotypes (Fig. 4). In the phenogram constructed based on the inoculation data, ten clusters were observed at the 50% similarity level. Among these clusters, cluster I, considered as major, comprises a maximum of 10 pathotypes out of 24, whereas clusters II, III, IV, VII and IX comprises double pathotypes. Clusters V, VI, VIII & X were represented by single pathotypes. From the phenogram, three significant patterns or relationship were observed: (i) groups of X. oryzae pv. oryzae isolates from different populations with same pathotypes showed multiple haplotypes (e.g., pathotype XII consisted of 7 distinct haplotypes), (ii), groups of X. oryzae pv. oryzae isolates from different populations had identical haplotype in multiple pathotypes (e.g., haplotype 1 is present in a maximum of 19 pathotypes), and (iii) groups of X. oryzae pv. oryzae isolates from the same population had identical haplotypes and pathotypes (haplotype 2 is identical for pathotype XXI). However, the strength of the relationship between pathotypes and the molecular haplotypes needs to be checked by comparing the similarity matrix of virulence with that of the PCR data.
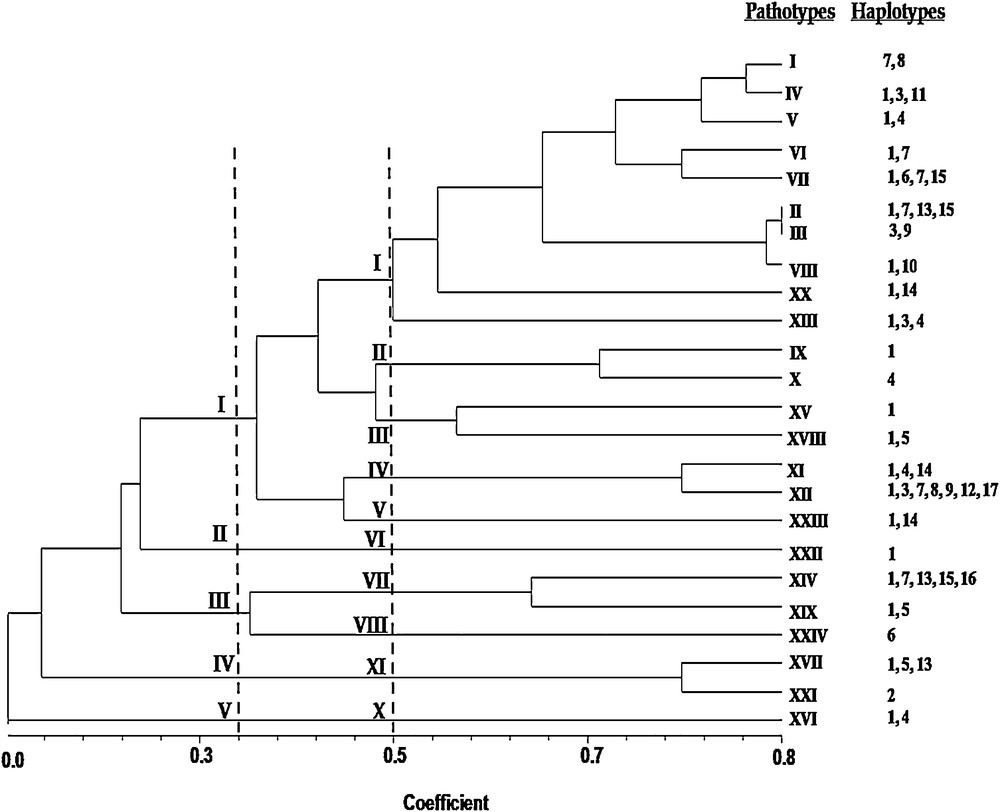
Phenogram constructed based on the virulence of X. oryzae pv. oryzae isolates to 12 near-isogenic lines containing a single gene for resistance showing the relationship between the pathotypes and the haplotypes. A data matrix was generated for the virulence data by scoring avirulence as 0 and virulence as 1. From these data, a phenogram was reproduced by the unweighted pair–group method for arithmetic average in the SHAN program (NTSYS-pc, version 1.80, Exeter Biological Software, Setauket, NY, USA). The dotted lines indicate the similarity index. I–X are the clusters of the pathotypes at the 50% similarity level. Masquer
Phenogram constructed based on the virulence of X. oryzae pv. oryzae isolates to 12 near-isogenic lines containing a single gene for resistance showing the relationship between the pathotypes and the haplotypes. A data matrix was generated for the virulence ... Lire la suite
4 Discussion
The identification of the pathogenic race of X. oryzae pv. oryzae is based on its ability to induce a combination of a compatible (susceptible) reaction and of an incompatible (resistant) one when inoculated on a standard set of differential host cultivars [1,14]. Near-isogenic rice lines (NILs) possessing different major genes for resistance to X. oryzae pv. oryzae were developed by IRRI and were used to assess the pathotypes of X. oryzae pv. oryzae worldwide. To assess the pathotypic and genetic variation of the BB pathogen, X. oryzae pv. oryzae in Bangladesh, a field survey was conducted in 19 rice-growing districts during the irrigated and rain-fed seasons in 2014. The analyses of the genetic diversity of X. oryzae pv. oryzae isolates using IS1112 marker resulted in a total of 17 molecular haplotypes, irrespective to the pathotypes. Most of the isolates used in this study were collected from a single rice variety, Swarna, (around 65%) in the rain-fed season, and from a hybrid one (around 75%) in the irrigated season, which were grown in different districts in Bangladesh during 2014. The prevalence of the same haplotypes in different rice-producing zones could be due to the widespread cultivation of Swarna and of the hybrid variety in different areas, and this cultivar may have served as a source of dissemination of the pathogen within the country. A similar study has also been conducted by Adhikari et al. [1]. They observed that most of the X. oryzae pv. oryzae strains (70%) tested in their study were collected from a single rice variety, Mansuli, which is grown in different locations in Nepal. On the basis of prevalence of the same haplotypes in different rice-producing zones, they assumed that it could be due to the widespread cultivation of Mansuli in different zones, and this cultivar may have served as a source of dissemination of the pathogen within Nepal. Although the sources of 96 strains in this study were different but host (rice cultivars) diversity had no effect on pathogen genetic diversity. Adhikari et al. [1] conducted an experiment in Nepal and reported that the diversities of X. oryzae pv. oryzae populations collected from traditional and improved rice cultivars were similar, suggesting that host diversity does not affect pathogen diversity. Based on a comparison of different agro-ecosystems and cultivars in the Philippines, Ardales et al. [4] also suggested that host diversity did not strongly affect pathogen diversity.
A dendrogram and a phenogram were constructed based on the high degree of DNA polymorphism of Xoo. All collected strains are pathogenically, genetically, and geographically diverse, showing a complex relationship between pathotypes and molecular haplotypes. Similar experimental evidences were demonstrated by Adhikari et al. [1], Nelson et al. [16], Ochiai et al. [18] and Yashitola et al. [22]. They stated that the relationship between molecular haplotypes and X. oryzae pv. oryzae races were complex.
A partial independent association between molecular haplotypes and pathotypes was observed from the dendrogram, as shown by the comparison of similarity matrices. For 65% similarity matrices, 17 molecular haplotypes/lineages under 10 clusters were identified from 96 Xoo isolates covering 19 separate geographic districts. This relationship appears to be due to a high degree of DNA polymorphism among all collected isolates under 24 pathotypes. Similar observations were found by Adhikari et al. [1]. They collected 171 strains from 8 rice-producing zones in Nepal and distinguished 31 molecular haplotypes using two polymerase chain reaction-based assays. Among them, some haplotypes were geographically dispersed and three haplotypes were detected as the most frequent ones, which occurred in diverse geographic populations. In the phenogram, 26 pathotypes grouped into five clusters, and cluster 1 contained strains with a wide virulence spectrum from all geographic populations. They reported two major groups of X. oryzae pv. oryzae in Nepal. Among them, one group performed high molecular polymorphism with representing maximum number of pathotypes.
From the constructed phenogram, three significant patterns were observed and described regarding the complex relationship between pathotype and molecular haplotype. Similar results were reported by Shanti et al. [20] and Adhikari et al. [1]. Shanti et al. [20] found that different isolates belonging to the same pathotype belonged to different haplotypes. Their study revealed that a wide pathogenic and genetic variation exists among the X. oryzae pv. oryzae isolates. On the other hand, Adhikari et al. [1] observed a weak correlation (r = 0.52) between molecular haplotypes and virulence phenotypes. They pointed out that, although the molecular variation was greatest between strains of different virulence phenotypes, some variation was observed among strains with identical virulence.
In the current study, genetically similar haplotypes were detected from different districts of Bangladesh, suggesting that there might be migration of X. oryzae pv. oryzae, perhaps due to national exchanges or regional movement of contaminated germplasm. George et al. [7] used PCR and RFLP markers to compare strains of X. oryzae pv. oryzae from Indonesia and the Philippines. They found that the predominant strains in the pathogen collections from both countries were closely related and concluded that there was a regional movement of the pathogen due to germplasm exchange.
Another important factor, the effects of host–pathogen interactions through gene-for-gene theory and change in the pathogenic structure through the mutation process responsible for the high degree of DNA polymorphism should be considered. Wolfe et al. [21], Eversmeyer and Browder [5] and Lebeda [11] solely evaluated the genetic relationships by either the frequency of virulence factors or quantified the degree of association between the resistance factor in the host and the virulence factor in the pathogen population, in a manner similar to the gene-for-gene relationship. Nayak [15] stated that the widespread repeated use of a few resistance genes might accelerate the selection of new pathogenic races at a rate equivalent to 1.64 times the increase in specific virulence of the isolates following the resistance host–plant selection pressure after a single crop cycle. This will lead to a change in the pathogen population structure through either mutation or recombination to adapt itself to the new resistant host plant or environmental changes.
5 Conclusion
This diversity may explain some of the difficulties encountered historically in developing rice cultivars with resistance to X. oryzae pv. oryzae in Bangladesh. The problems of understanding the biology of X. oryzae pv. oryzae and controlling BB have long been challenging. The known geographic distribution of the haplotypes provides a baseline for monitoring the future spread of X. oryzae pv. oryzae in other rice producing areas in Bangladesh. The present study may serve as a platform for refined characterization of new isolates whose racial classification has not been determined. More isolates of all rice-growing areas are needed to be analyzed in order to fully understand the population structure of X. oryzae pv. oryzae existing in Bangladesh. The results of this study and further DNA fingerprinting analyses would be useful in the selection of strains for additional resistance screening.
Disclosure of interest
The authors declare that they have no competing interest.
Acknowledgement
This work was supported by the International Foundation for Science (IFS) (Grant No. C/5035-1) awarded to Dr. Md. Rashidul Islam, Department of Plant Pathology, BAU). The authors would like to acknowledge the Bangladesh Rice Research Institute (BRRI) for providing the IRBB NILs.



Vous devez vous connecter pour continuer.
S'authentifier