1 Introduction
Capparis spinosa L. (the thorny caper, Capparaceae) is a xerophyte plant mainly exploited as a condiment to flavor foods [1–3]. It has a wide geographic distribution in the Mediterranean area and in Central Asia [4,5], and is adapted to several bioclimatic environments, from humid to Saharan [4,6–8] and to different soil types [4,8–10]. It has a high phenotypic diversity and several morphological distinct intraspecific taxa and intermediate forms were reported making its classification very ambiguous [10–13]. In addition, morphological markers are influenced by the environmental conditions and developmental stages, which make their use limited in diversity studies. It has become obvious these days that morphological characterization of the species of Capparis is not sufficient to make definitive discrimination among the species, subspecies, and varieties [14]. More recently, alternate approaches, including application of appropriate molecular markers, have been increasingly adopted to address the problems in Capparis taxonomy. Analyses on genetic diversity and relationship among the species of Capparis could also provide useful information for the conservation of genetic resources and the establishment of a Capparis breeding program. In fact, several recent works assessed the biological activities of caper extracts (whole plant, floral roots, sheets, buds, flowers and fruits) on bacteria [15–20], fungi [19,21], nematodes [22], pests [23] and adventitious [24], and suggested that C. spinosa is a potential source of natural antioxidant molecules [3,25–27]. Genetic diversity studies could give a general guide for choosing parental lines to make suitable cross combinations for the selection of valuable traits with large possible applications in agriculture, food industry and medicine.
The inter-simple sequence repeats (ISSRs) and random amplified polymorphic DNAs (RAPDs) analysis have been the most commonly used techniques in wild and cultivated forms of Capparis species, revealing genetic variation among genotypes/species and/or collected sites in Iran [28], Trans-Himalayan region [29], Syria [30], Turkey [31], Egypt [32], Italy [33] and Morocco [34]. In Syria, ISSR combined with simple sequence repeat (IRAP) revealed genetic variation among 47 samples of three Capparis species genotypes collected from 21 locations and divided the samples into three genetic groups: Capparis sicula, C. aegyptia and C. spinosa [30]. In Morocco, ISSR markers identified 20 genetic groups among 90 accessions of Capparis spp. [34]. In Italy, Gristina et al. [33] characterized 90 wild populations and cultivated forms using ISSR markers and suggested a clear genetic distinctness between two different subspecies of C. spinosa at the regional level, C. spinosa subsp. spinosa and subsp. rupestris. However, another recent study on eight Egyptian taxa of Capparis and related genera (Capparaceae) conducted by Moubasher et al. [32] using three primers in randomly amplified polymorphic DNA (RAPD) analyses revealed four varieties of Capparis spinosa: C. spinosa var. deserti, C. spinosa var. canescens, C. spinosa var. spinosa and C. spinosa var. inermis. Using RAPD technique, Özbek and Kara [31] also differentiate five different varieties, Capparis spinosa L. var. spinosa, var. aegyptia and var. canescens and C. ovata Desf. var. palaestina, and var. herbacea, and intermediate forms between 15 Turkish natural Capparis populations. In the other hand, among the different molecular tools, the Amplified Fragment Length Polymorphism (AFLP) method has been extensively used for a wide range of species, including medicinal plants [35–37], to investigate the population genetic structure and assess the genetic differentiation among species [38,39]. This technique is a powerful DNA fingerprinting technology applicable to any organism without the need for prior sequence knowledge. It is a reproducible multilocus marker system with selective PCR amplification [40]. The main advantages of AFLPs are the high levels of polymorphism and high degrees of discriminative capacity for closely related accessions [41–45]. This technique was used successfully in C. spinosa, revealing genetic variations among 28 samples collected from six sites in the Aleppo and Lattakia provinces (Syria), and showed the presence of specific alleles for each province [46].
In Tunisia, C. spinosa is widely distributed. Although it has been extensively studied morphologically, Capparis taxonomy is also still highly controversial. Pottier-Alapetite [47] was the first to state four varieties (var. canescens, var. rupestris, var. genuina and, var. aegyptiaca) based on morphological traits. Later, Le Floc’h et al. [48], based on Inocencio et al.’s [8] and Yousfi's [49] phenotypic studies, sustained six Capparis species in Tunisia: C. spinosa subsp. spinosa var. spinosa, C. sicula subsp. sicula, C. orientalis, C. ovata subsp. ovata, C. aegyptia, and C. zoharyi, a new species mentioned for the first time. More recently, Fici's [10] works, based on morphological, ecological, biogeographical characteristics and on herbarium samples, suggested another classification and grouped intraspecific taxa of C. spinosa into two subspecies in Tunisia based on the presence of persistent thorns: C. spinosa subsp. spinosa, the thorny morphotype, with three varieties (var. canescens, var. spinosa, and var. aegyptia) and C. spinosa subsp. rupestris, the inerm morphotype, with two varieties (var. ovata and var. rupestris). According to Fici [10], C. zoharyi is not morphologically distinct from C. aegyptia. Until now genetic studies on Tunisian Capparis were rare and limited to one variety/species or one region, not helping to resolve this unclear classification. Khouildi et al. [50] was the first to study genetically Capparis species in Tunisia and showed a higher variability in Tunisian caper populations compared to Italian populations using RAPD analysis. In addition, Ghorbel et al. [51] subdivided 12 Tunisian Capparis populations into two genetic groups belonging to inerm and thorny morphotypes on the basis of RAPDs and isoenzymatic tools.
The objective of this study were:
- • to molecularly characterize a representative panel of the morphologically identified Capparis accessions in Tunisia by AFLP analysis;
- • to study the population structure, the phylogenetic relationship between accessions, and gene flow;
- • to study the correlation of the genetic diversity with geographical and ecological suitability of Capparis in Tunisia.
2 Material and methods
2.1 Plant materials and DNA extraction
Two hundred and thirteen caper accessions, sampled from different climate zones, were selected for this study from the collection conserved at the Tunisian National Gene Bank (see details in supplementary material). Accessions belong to six distinct caper species (C. sicula, C. spinosa, C. ovata, C. orientalis, C. zoharyi, and C. aegyptia) and were chosen based on clear morphological characterizations [48]. Sampling site and GPS coordinates were recorded for each accession.
Total genomic DNA was extracted from frozen young leaves according to a modified CTAB procedure described by Saghai-Maroof et al. [52]. DNA concentration was estimated by spectrophotometer and by electrophoresis on [1% (w/v)] agarose gel [53].
2.2 AFLP genotyping
For each accession, 10 μL of genomic DNA (500 ng) was double digested with 5U of EcoRІ restriction enzyme at 37 °C in a final volume of 20 μL for 4 h, then with 5U of MseІ restriction enzyme at 65 °C in a final volume of 40 μl for 4 hours. The resulting fragments were ligated to double-stranded EcoRI (5 pmol) and MseI (50 pmol) in ligation buffer (4UT4 DNA ligase, 10X T4 DNA ligase buffer) and incubated at 37 °C for 16 h. Ligated DNA templates were further diluted (5 fold) and pre-amplification was performed using EcoRІ+ A and MseІ+ C primers in a total volume of 25 μL (0.20 μL of Taq, 1 μL of enzyme, 0.2 μL of dNTP, 1.5 μL of MgCl2 and 2.5 μL of buffer). The pre-amplification reaction was carried out in a thermocycler for 2 min at 94 °C, 30 cycles at 94 °C denaturation (30 s), 56 °C annealing (30 s) and 72 °C extension (1 min) and a final hold at 72 °C for 10 min. Pre-amplified DNA was analyzed by 1%agarose gel electrophoresis. The pre-selective amplification product was diluted 25× in a TBE buffer 1X and stored at 4 °C for amplification, or stored at −20 °C for later use. Selective amplification was performed in a final volume of 20 μL containing 1.5 μL of buffer, 0.2 μL of dNTP, 1.25 μL of MgCl2, 0.2 μL of Taq, 5 ng of EcoRI+ AXX and 30 ng of MseI + CYY and 5 μL of a diluted pre-amplified DNA sample. Amplifications were conducted using a touchdown PCR program: 1 cycle of 94 °C for 30 s, 65 °C for 30 s, and 72 °C for 60 s, then 13 cycles with the annealing temperature lowered by 0.7 °C per cycle, followed by 23 cycles of 94 °C for 30 s, 56 °C for 30 s, and 72 °C for 60 s. One microliter of the amplified product was mixed with 13.5 μL of deionized formamide and 0.5 μL of GeneScan-500 Liz internal size standard, denaturized at 95 °C for 5 min and analyzed by capillary electrophoresis on an automated ABI Prism 3130 DNA sequencer.
A screening of 12 combinations of four EcoRI primers (E-AAC, E-ACC, E-ACA and E-AAG) and six MseI primers (M-CTC, M-CAG, M-CAT, M-AGG, M-GCT and M-CAA) with three selective nucleotides were performed. Three highly polymorphic primer combinations were selected: E-AAC/M-CAG, E-AAG/M-CAA and E-ACC/M-CAT (Table 1).
Number of AFLP bands, percentage of polymorphic bands, polymorphism information content (PIC) and resolving power (Rp) for each AFLP primer combination used on 213 Capparis accessions from Tunisia.
| Primer pairs | Total AFLP bands (No.) |
Polymorphic AFLP bands (No.) |
Percent Polymorphism (%) |
PIC | Rp | Fragment size range (bp) |
| E-AAC/M-CAG | 262 | 203 | 77.48 | 0.242 | 67.06 | 50–477 |
| E-AAG/M-CAA | 233 | 210 | 90.12 | 0.254 | 74.25 | 53–480 |
| E-ACC/M-CAT | 255 | 223 | 87.45 | 0.262 | 81.9 | 57–497 |
| Total | 750 | 636 | – | – | 223.21 | – |
| Average | 250 | 212 | 85.02 | 0.252 | 74.40 |
2.3 AFLP polymorphism analysis
Clear and unambiguous bands in length ranging from 50 to 500 pb were considered as usable. AFLP bands were scored, across all genotypes, for presence (1) or absent (0) and transformed into 0/1 binary matrix. The total number of bands was calculated for all primers. Polymorphic bands were only taken into account to estimate the percentage of polymorphic bands (%PB). The ability of the most informative primers to discriminate among cultivars was assessed by the resolving power (Rp) [54]. Evaluation of the Rp was performed according to the formula of Gilbert et al. [55]. In addition, the discriminating power of derived markers was made by the assessment of the polymorphism information content (PIC) using Lynch and Walsh [56] formula.
2.4 Population structure analysis
Population genetic structure was assessed using the Bayesian clustering algorithm implemented in STRUCTURE version 2.3 [57,58]. The program infers the number of populations into which the analyzed genotypes can be divided. The samples were analyzed assuming the admixture ancestry model with K from 1 to 10 applying 4 independent runs for each of the different values of K. A burn-in period of 50,000 and Monte Carlo Markov Chain replicates of 100,000 iterations were performed. The run with maximum likelihood was used to assign individual genotypes into groups. Within a group, genotypes with affiliation probabilities (inferred ancestry) ≥ 80% were assigned to a distinct group and those with < 80% were treated as “admixture”. Plot of mean posterior probability (ln P(D)) values per clusters (K), generated by the STRUCTURE program [57] and delta-K method of ln P(D) [59] were used to determine the optimal number of clusters. Principal coordinate analysis (PCoA) was also performed via distance matrix to describe the relationship between accessions using software GenAlEx version 6.5 [60].
2.5 Genetic diversity, genetic distances and phylogeny
The significance of population differentiation clustered by STRUCTURE was further investigated by performing an analysis of molecular variance (AMOVA). The gene flow was estimated by using the equation
2.6 Neutrality test
The Tajima's D test is a widely used test of neutrality in population genetics to assess the random or non-random evolutionary process in a species [63]. Tajima's D [63] test was performed for each population individually using Mega 6 software. Tajima's statistic computes a standardized measure of the total number of segregating sites in the sampled population and the average number of polymorphic sites between pairs in a sample. The null hypothesis of the Tajima's D test is neutral evolution in an equilibrium population. The theoretical distribution of Tajima's D (95% confidence interval between −2 and +2) assumes that polymorphism ascertainment is independent of allele frequency. Positive values of Tajima's D suggest an excess of common alleles, which can be consistent with balancing selection, population contraction or bottleneck; whereas negative results are indicative of a population that has undergone a recent expansion, as rare alleles are more common than expected.
3 Results
3.1 AFLP polymorphism
Three primer combinations generated a total of 750 reproducible amplification fragments across all caper accessions among which 636 were polymorphic. The average number of polymorphic bands scored per primer pair was 210 (Table 1). All the tested primers were powerful to detect DNA polymorphisms in Capparis genus with a similar level of polymorphism. The largest number of polymorphic bands 223 was produced with primer combination E-ACC/M-CAT and the least number of polymorphic bands 203 was detected using primer combination E-AAC/M-CAG. Moreover, estimates of the resolving power (Rp) showed a high rate of collective Rp (223.21) with an average of 74.4. The Polymorphism Information Content (PIC) ranged from 0.242 to 0.262, with an average of 0.252 per primer pair. The most informative primer combination for distinguishing the genotypes was E-ACC/M-CAT, with the highest values of Rp (81.9) and PIC (0.262) (Table 1).
3.2 Population structure
The program STRUCTURE [57,58] was run independently four times, with K ranging from 1 to 10, in order to study the structure of the caper population analyzed. The plot of mean posterior probability (ln P(D)) values per clusters (K) as well as the ΔK plot based on ln P(D) values indicated that the most likely number of populations (K) was 6 (Fig. 1A and B). Each morphological group of caper, C. spinosa subsp. spinosa var. spinosa, C. sicula subsp. sicula, C. orientalis, C. ovata subsp. ovata, C. aegyptia and C. zoharyi, was distinct and clustered into a separate genetic group named c.si, c.sp, c.or, c.ov, c.ae, and c.zo, respectively. The graphic representation of the estimated membership coefficients to the clusters for each accession (at K = 6) is shown on Fig. 2. At K = 2, C. sicula subsp. sicula and C. spinosa subsp. spinosa var. spinosa represented by c.si and c.sp respectively, split off from the other species and together formed a separate subpopulation. Increasing K to 3, the accessions largely separated into a C. sicula subsp. sicula–C. spinosa subsp. spinosa var. spinosa group, a C. orientalis–C. ovata subsp. ovata group and a group containing C. aegyptia–C. zoharyi. The population structure was also assessed individually for each AFLP primer combinations leading to the same clustering (data not shown). Thirty percent of admixed individual was observed in the population; 80% of them were morphologically identified as C. ovata subsp. ovata. These hybrids are mixed between C. sicula subsp. sicula, C. spinosa subsp. spinosa var. spinosa and C. orientalis at about 33% each.

Plot of mean posterior probability (ln P(D)) values (A) per cluster (K), based on 4 replicates per K, for K ranging from 1 to 10, generated by the STRUCTURE program [57], and delta-K analysis (B) of ln P(D), according to Evanno et al. [59]. A burn-in period of 50,000 and Monte Carlo Markov Chain replicates of 100,000 iterations were used.

Proportion of membership coefficient at K = 6 for 213 Tunisian Capparis accessions belonging to 6 species using 636 amplified fragment length polymorphism (AFLP) loci under STRUCTURE software. The different genetic populations, c1, c2, c3, c4, c5 and c6, identified under STRUCTURE were color-coded in light blue, pink, yellow, dark blue, red, and green, respectively. Genotypes are ordered by their morphological similarities to a particular Capparis species, c.si, c.sp, c.or, c.ov, c.ae and c.zo correspond to C. sicula subsp. sicula, C. spinosa subsp. spinosa var. spinosa, C. orientalis, C. ovata subsp. ovata, C. aegyptia, and C. zoharyi species, respectively. Masquer
Proportion of membership coefficient at K = 6 for 213 Tunisian Capparis accessions belonging to 6 species using 636 amplified fragment length polymorphism (AFLP) loci under STRUCTURE software. The different genetic populations, c1, c2, c3, c4, c5 and c6, identified ... Lire la suite
The PCoA of the 213 caper accessions showed that the first two axes accounted respectively for 16.19% and 15.19% of the genetic variation, explaining altogether 31.38% of the total variation. The second axis clearly separated the three clusters G1, G2 and G3, while the first axis differentiates the species within clusters G1 and G2. Color-coding of the accessions in the 2-dimensional PCoA plot showed a good correspondence with the population groups obtained from the STRUCTURE analyses (Fig. 3). The presence of intermediate genotypes between clusters confirmed STRUCTURE results of admixed individuals (Fig. 2).

Principal coordinate plot depicting the six genetic populations of Capparis within 213 Tunisian accessions using 636 amplified fragment length polymorphism (AFLP) loci under GenAlex software. The different genetic populations, c1 to c6, identified under STRUCTURE were color-coded in light blue, pink, yellow, dark blue, red, and green, respectively. Genotypes were color-coded according to their membership to each of the six genetic populations. hy, color-coded in gray, referred to admixed individuals according to STRUCTURE results.
3.3 Analyses of molecular variance and diversity indices
The six populations identified from STRUCTURE analysis were also subjected to analysis of variance (AMOVA) to estimate the percentage of variation among populations and within population. The AMOVA indicated a highly significant (P < 0.01) genetic differentiation within the sampled Capparis populations. Fifty percent of the total genetic variance was attributed to the populations based on structure, and the remaining 50% was explained by individual differences within population (Table 2a). In addition, ours results determined a high number of specific alleles for each species (population) (Table 3). Capparis spinosa subsp. spinosa var. spinosa presented the highest number of specific alleles (103 AFLP loci) and C. ovata subsp. ovata presented the smallest number of specific alleles (42 AFLP loci). This result indicated that C. spinosa subsp. spinosa var. spinosa and C. ovata subsp. ovata were probably the most differentiated and non-differentiated populations, respectively. We also showed that C. ovata subsp. ovata has the less genetic deviation between accessions (d = 0.071), while C. orientalis has the highest deviation indices (d = 0.137). This indicates that C. ovata subsp. ovata is the less diverse population, while C. orientalis is the most diverse (Table 3).
Analysis of molecular variance (AMOVA) of 213 Tunisian Capparis accessions belonging to six genetic populations (a) and 4 bioclimatic stages (b) using 636 AFLP loci performed under GenAlex.
| Source of variation | Degree of freedom | Sum of squares | Variance component | Estimated variation | Percentage of variation | P-value |
| a. Genetic populations | ||||||
| Among populations | 5 | 10742.230 | 2148.446 | 61.98 | 50 | 0.01 |
| Within population | 207 | 12724.624 | 61.47 | 61.47 | 50 | |
| Total | 212 | 23466.854 | 123.45 | 100 | ||
| b. Bioclimatic stages | ||||||
| Among bioclimatic stages | 3 | 5573.85 | 1857.95 | 37.91 | 31 | 0.01 |
| Within bioclimatic stage | 209 | 17892.99 | 85.61 | 85.61 | 69 | |
| Total | 212 | 23466.85 | 123.45 | 100 |
Diversity indexes within each Tunisian Capparis populations and Tajima's Neutrality Test calculated using MEGA software.
| Genetic codea | M | S | p s | Θ | Π | D (P-value) | Specific AFLP bands (number: size – range in bp) | d |
| c.si | 12 | 219 | 0.232 | 0.077 | 0.081 | 0.298 (0.081) | 62: 69 – 497 | 0.082 |
| c.sp | 45 | 597 | 0.634 | 0.145 | 0.131 | −0.334 (0.131) | 103: 55 – 446 | 0.132 |
| c.or | 60 | 605 | 0.642 | 0.137 | 0.136 | −0.024 (0.136) | 53: 51 – 499 | 0.137 |
| c.ov | 8 | 157 | 0.166 | 0.064 | 0.070 | 0.562 (0.070) | 42: 67 – 480 | 0.071 |
| c.ae | 25 | 466 | 0.495 | 0.131 | 0.107 | −0.728 (0.107) | 82: 70 – 492 | 0.107 |
| c.zo | 37 | 519 | 0.551 | 0.132 | 0.122 | −0.273 (0.122) | 65: 62 – 406 | 0.123 |
a c.si, c.sp, c.or, c.ov, c.ae and c.zo were defined according to STRUCTURE results and correspond to the morphologically distinct species C. sicula subsp. sicula, C. spinosa subsp. spinosa var. spinosa, C. orientalis, C. ovata subsp. ovata, C. aegyptia and C. zoharyi, respectively.
3.4 Phylogenetic analysis and genetic distances
The dendrogram constructed using UPGMA clustering method separated all the accessions of six species of caper studied into three main clusters; and each species constitute a unique subgroup as described in STRUCTURE analysis (Fig. 4). The first cluster, marked G1, grouped the two species C. sicula subsp. sicula (c1) and C. spinosa subsp. spinosa var. spinosa (c2). The second cluster G2 contained the two species C. orientalis (c3) and C. ovata subsp. ovata (c4). The third cluster G3 grouped the two species C. aegyptia (c5) and C. zoharyi (c6). In each subgroup (species), all genotypes shared the same ancestor.
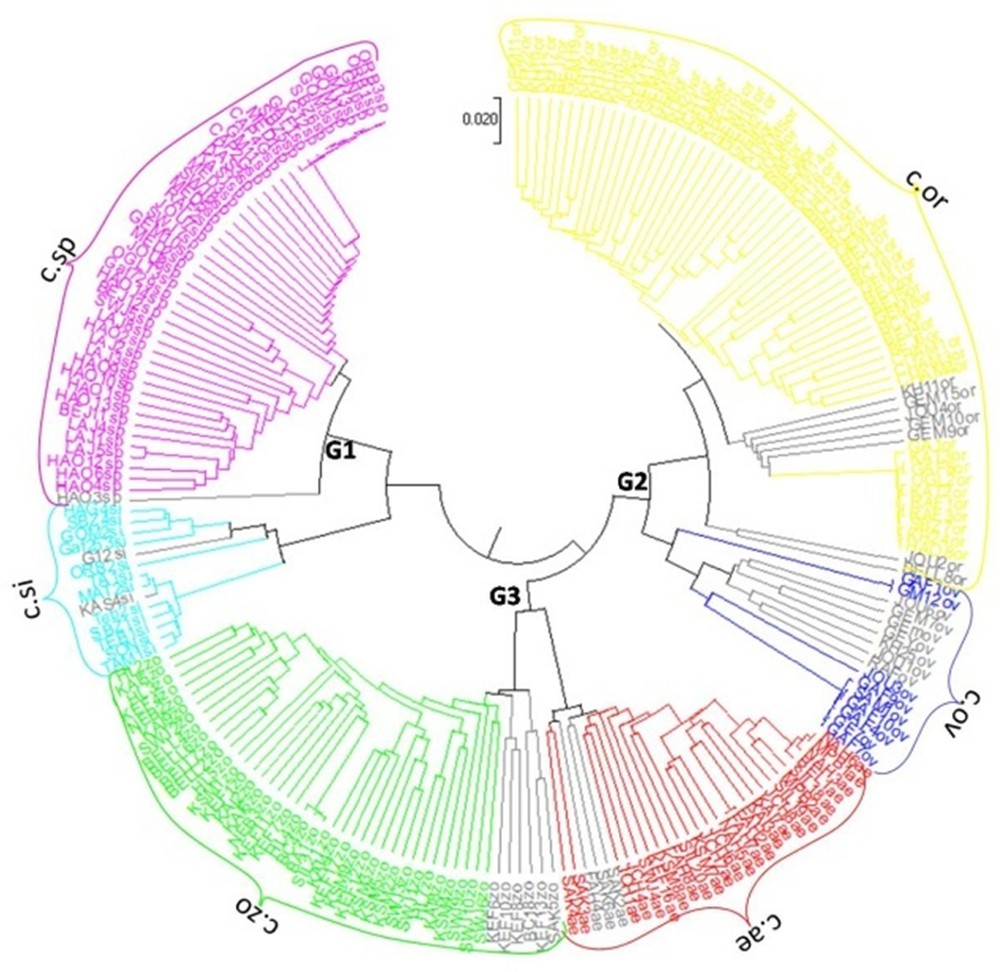
UPGMA clustering of Nei's genetic distance based on the amplified fragment length polymorphism (AFLP) data of 213 Tunisian Capparis accessions performed under Mega software. Genotypes were color-coded in light blue, pink, yellow, dark blue, red and green according to their membership to c1, c2, c3, c4, c5 and c6 respectively. Admixed individuals were color-coded in gray. G1, G2 and, G3 correspond to the three main genetic clusters identified. c.si, c.sp, c.or, c.ov, c.ae and c.zo correspond to the morphologically distinct species of C. sicula subsp. sicula, C. spinosa subsp. spinosa var. spinosa, C. orientalis, C. ovata subsp. ovata, C. aegyptia and, C. zoharyi, respectively. Masquer
UPGMA clustering of Nei's genetic distance based on the amplified fragment length polymorphism (AFLP) data of 213 Tunisian Capparis accessions performed under Mega software. Genotypes were color-coded in light blue, pink, yellow, dark blue, red and green according to ... Lire la suite
Genetic similarity coefficients of the six genetic groups (species) based on Nei's genetic distance is given in Table 4. The high values of genetic similarity coefficients indicated close genetic relationships between populations and the low values indicated remote relationships among populations. The highest similarity value (0.298) was recorded between C. spinosa subsp. spinosa var. spinosa and C. aegyptia, while the second highest similarity value (0.286) was found between C. spinosa subsp. spinosa var. spinosa and C. zoharyi. In addition, the lowest Nei's genetic distance (0.223) was found between C. zoharyi and C. ovata subsp. ovata, and between C. aegyptia and C. zoharyi, indicating that C. zoharyi and C. ovata subsp. ovata, as well as C. zoharyi and C. aegyptia, are the most genetically related species. Similarly, low Nei's distances were also recorded between C. sicula subsp. sicula and C. spinosa subsp. spinosa var. spinosa, and between C. orientalis and C. ovata subsp. ovata with 0.225 and 0.234 respectively. Furthermore, pairwise Nm values ranged from 0.226 to 0.588 suggesting frequent gene flow among populations. Similar levels of gene flow were recorded between populations within genetic cluster as Nm values between C. sicula subsp. sicula and C. spinosa subsp. spinosa var. spinosa, between C. orientalis and C. ovata subsp. ovata, and between C. aegyptia and C. zoharyi were 0.522, 0.504 and 0.588, respectively (Table 4).
Pairwise Nei's genetic distances (lower diagonal) and Nm values (upper diagonal) between the Capparis populations based on AFLP data.
| c.si | c.sp | c.or | c.ov | c.ae | c.zo | |
| 0.000 | 0.522 | 0.443 | 0.260 | 0.288 | 0.352 | c.si |
| 0.225 | 0.000 | 0.483 | 0.428 | 0.350 | 0.405 | c.sp |
| 0.253 | 0.273 | 0.000 | 0.504 | 0.408 | 0.502 | c.or |
| 0.226 | 0.249 | 0.234 | 0.000 | 0.374 | 0.476 | c.ov |
| 0.270 | 0.298 | 0.275 | 0.225 | 0.000 | 0.588 | c.ae |
| 0.267 | 0.286 | 0.258 | 0.223 | 0.223 | 0.000 | c.zo |
3.5 Genetic diversity is correlated with geographic distance
Mantel's test in the entire Capparis population rejected the null hypothesis as a correlation (Rxy = 0.145; P < 0.001) between genetic and geographic distances was observed among accessions. However, this was not true when the Mantel test was performed for accessions within each cluster individually suggesting that each population is geographic localized (Table 5). Population clustering largely corresponded to climatic zones in Tunisia (Fig. 5). The two species of the G1 cluster, C. spinosa subsp. spinosa var. spinosa and C. sicula subsp. sicula were mostly (at 83.33%) grown in Saharan bioclimate; the two species of the G2 cluster, C. orientalis and C. ovata subsp. ovata, were frequent at 70% in arid bioclimate; while 95% of the two species of G3 cluster, C. aegyptia and C. zoharyi, were grown in semi-arid bioclimate. In addition, the major frequencies of hybrid species (92%) were present in northern Tunisia, represented by the subhumid and semi-arid bioclimates. The subhumid bioclimate grouped the highest number of species (5 out of 6). Capparis spinosa subsp. spinosa var. spinosa and C. orientalis were the only two species identified in all bioclimatic stages. The analyses showed a significant correlation (r = 0.956; P < 0.0001) between the number of hybrids and the diversity indices of the bioclimatic stages. A north–south gradient revealed the highest number of hybrids, and diversity indices were observed in northern Tunisia (represented by the subhumid and semi-arid bioclimates), whereas the genetic diversity drops toward the south (represented by the arid and Saharan bioclimates) (Fig. 6). Our AMOVA analysis revealed that 31% of the total genetic diversity is explained by the bioclimatic stages (Table 2b).
Correlation between genetic distance and geographic distance for each of the genetic cluster G1, G2 and G3 of Capparis in Tunisia based on a Mantel's test using AFLP data.
| Genetic clustera | R xy | P(Rxy-rand ≥ Rxy-data) |
| All | 0.145 | 0.001 |
| G1 | 0.177 | 0.01 |
| G2 | 0.139 | 0.02 |
| G3 | 0.055 | 0.06 |
a Genetic clusters G1 to G3 were defined by UPGMA analysis.

Geographic structure of 213 Capparis accessions over the Tunisian climatic zones showing the differential localization of the six genetic populations as defined by STRUCTURE. The six different genetic populations c1 to c6 were color-coded in light blue, pink, yellow, dark blue, red, and green, respectively. hy, color-coded in gray, referred to admixed individuals according to STRUCTURE results.
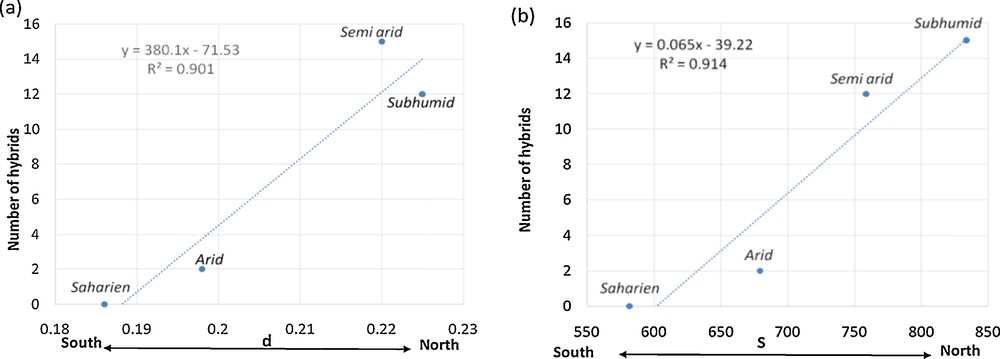
Correlation between the number of hybrids and the estimate of average evolutionary divergence over amplified fragment length polymorphism (AFLP) fingerprint pairs in same group (a) and the number of segregating AFLP bands across the bioclimatic stages (b). d: the estimate of average evolutionary divergence over AFLP fingerprint pairs in same group; S: number of segregating AFLP bands.
3.6 Neutrality test
Tajima's D [63] test was performed to identify populations that do not fit the neutral theory model. The neutrality test of Tunisian Capparis sp. displayed significant negative Tajima's D values for five populations, indicating an excess of rare variation, consistent with population growth (Table 3). The population of C. aegyptia presented the highest negative value of Tajima's D (−0.728), while for C. orientalis Tajima's D value was the closest to zero (−0.024). For C. sicula subsp. sicula and C. ovata subsp. ovata, Tajima's D value was positive, indicating balancing selection.
4 Discussion
Caper classification is ambiguous and highly controversial; its population structure has been poorly studied. Various taxonomists have recognized 250 morphologically different species in the genus Capparis. Most of the confusion is due to free hybridization of different species and occurrences of intermediate forms. The perplexing condition of the Capparis phylogeny has driven many researchers to resolve the classification ambiguities of wild and cultivated forms of C. spinosa genotypes using molecular markers. The objective of this study was to genetically characterize 213 Capparis sp. accessions sampled from diverse sites in Tunisia in order to study population structure and to correlate the genetic makeup with the morphological description and geographical distribution of Capparis species.
The three AFLP primer combinations revealed the same level of polymorphism and diversity indices, and showed the same power to discriminate between the Capparis species. Our results support the importance of AFLP analysis where only one primer combination is sufficient for diversity studies. In the literature, very scarce information regarding the genetic diversity of Capparis species is known. Our AFLP results revealed a high polymorphism level in the Tunisian Capparis genus similarly to previous RAPD [28,29,31,32], and ISSR [29,33,34] studies were the polymorphism among Capparis accessions ranged from 48.8% to 98% and from 75.51% to 100%, respectively, with an average of 12.3 polymorphic fragments per primer. The previous AFLP analysis in Capparis was performed in 45 accessions from Spain, Morocco, and Syria, and revealed 50% of polymorphic bands and showed that C. aegyptia and C. ovata are quite separated from the other taxa. The two groups correspond to the populations from Syria as opposed to Morocco and Spain, showing that geographical differences exist [64]. Also, this analysis was performed between samples collected from six sites in Aleppo and Lattakia provinces (Syria) and revealed a certain number of specific alleles for each provinces and the presence of genetic and reproductive isolation deterrent to gene flow between the two provinces [46]. However, the comparison of our AFLP diversity level in Tunisian Capparis populations with previous genetic studies worldwide is difficult. The amount of genetic diversity is related to the marker used and to the Capparis population and its size [64].
The present molecular study helped to solve taxonomical ambiguities and the genetic relationship among caper accessions in Tunisia. Out of the 636 polymorphic fragments generated, 407 were species-specific. Our STRUCTURE study was able to distinguish six populations of Capparis in Tunisia with their AFLP fingerprint, corresponding to the six morphologically characterized species C. orientalis, C. ovata subsp. ovata, C. aegyptia, C. zoharyi, C. sicula subsp. sicula and C. spinosa subsp. spinosa var. spinosa. This genetic result was in line with the morphological classification of Inocencio et al. [8], Le Floc’h et al. [48], and Yousfi [49] The results obtained in this study both at the individual and population levels indicated that C. zoharyi constitutes a separate gene pool from the other species, especially from C. aegyptia and should be considered as a species by itself, contradicting with Fici [10] morphological classification. In our study, C. zoharyi presents 65 specific alleles and is genetically as distant from C. aegyptia as C. sicula subsp. sicula is distant from C. spinosa subsp. spinosa var. spinosa and C. ovata subsp. ovata is distant from C. orientalis. In addition, our phylogenetic analysis clarified the relationship between the six species and suggested their classification into three main genetic clusters. The first cluster (G1) grouped the two species C. spinosa subsp. spinosa var. spinosa and C. sicula subsp. sicula. These species have a procumbent habit, with persistent stipules that are not thorny, but are slender and curved, and grow mainly in the Saharan bioclimate. The second cluster (G2) contained the two species C. orientalis and C. ovata subsp. ovata, having a pendulous habit, deciduous stipules that are not thorny, but are slender and straight, and growing mainly in the subhumid bioclimate. The third cluster (G3) grouped the two species C. aegyptia and C. zoharyi that have an erect habit, with evergreen stipules that are spiny, wide and crooked and growing mainly in the semi-arid bioclimate. The geographic distribution pattern of these species in Tunisia and the spatial proximity between species within genetic clusters probably explain the similar gene flow level found in our study between C. sicula subsp. sicula and C. spinosa subsp. spinosa var. spinosa, between C. orientalis and C. ovata subsp. ovata, and between C. aegyptia and C. zoharyi within G1, G2 and G3 genetic clusters, respectively. In addition, this study revealed that accessions from G2 and G3 are more genetically related compared to accessions from G1. These genetic results refute again Fici's [10] morphological classification where C. aegyptia (G3here) was shown to be more closely related to C. spinosa subsp. spinosa var. spinosa and C. sicula subsp. sicula (G1), because of its persistent thorns, than to C. orientalis and C. ovata subsp. ovata (G2 here). Our results also contradict the previous work by Saadaoui et al. [65] reporting that C. spinosa subsp. spinosa was found in northern Tunisia (subhumid and semi-arid bioclimatic zones), whereas subsp. rupestris (C. orientalis) had a larger geographical distribution from subhumid to Saharan regions. On the other hand, the present study revealed that C. spinosa is commonly grown in the wild in diverse geographical and climatic regions in Tunisia contradicting Inocencio et al. [8] studies reporting that most known populations of C. spinosa were cultivated. In addition, our study did not support Inocencio et al. [64] hypothesis on the hybrid origin of C. spinosa between C. orientalis and C. sicula. Previous studies considering C. aegyptia as a varietal rank of C. spinosa (C. spinosa var. aegyptia (Lam.) Boiss) [4,61,66] were also refuted here.
This study was the first to cover most of the geographical and ecological areas and the various Capparis species growing in Tunisia. The AFLP polymorphism found between species suggests high variations at the genomic level that might have been accumulated during speciation events in Capparis as is evident from its morphological diversity. In addition, genotypes within species were genetically diverse; 50% of the total genetic diversity is observed within species. According to Khouildi et al.’s [50] results, the genetic variation found could be more likely related to environmental factors. Our analysis revealed that 31% of the total genetic diversity is explained by the bioclimatic stages. Ecological adaptations of Capparis to different environments could have been driven by isolation. The particularly high level of genetic variation, the negative value of Tajima's D within species, the population structure, and the presence of geographical patterns in diversity suggest several hypotheses regarding the population history of Capparis species. First, Capparis did not recently colonized Tunisia, and we confirm here its endemicity. Second, Capparis species may be undergoing population expansion and may have large population sizes. Third, our results revealed intermediate genetic diversity levels in C. aegyptia and C. zoharyi while C. sicula subsp. sicula and C. ovata subsp. ovata had low diversity indices. Assuming a same evolutionary rate in all species, we can suggest that through speciation, the divergence between C. aegyptia and C. zoharyi occurred earlier than the divergence between C. sicula subsp. sicula and C. spinosa subsp. spinosa var. spinosa, and the divergence between C. orientalis and C. ovata subsp. ovata. Fourth, Capparis's first settlement occurred in Northern Tunisia and then migrated during speciation across the country and colonized the southern regions. Subhumid bioclimate of Northern Tunisia is the most diversified grouping the three genetic clusters and five species out of six and is probably similar to the origin climate of the ancestral form. This genetic study also revealed that hybridization events are more frequent in the northern area, where all the genetic clusters are in sympatry. The region of greatest diversity was shown to be the origin area for many other wild species in plants [67–70]. Furthermore, from our phylogenetic results, we suggest C. spinosa subsp. spinosa var. spinosa (G1 genetic cluster), a species reported in all bioclimate (from subhumid to Saharan) and with high diversity indices, as the closest species to the ancestral form that generated the different Capparis species. Capparis spinosa subsp. spinosa var. spinosa was probably the first species to be differentiated during speciation in Capparis and had enough time to evolve and adapt to different geographic areas. In fact, Renfew [71] mentioned that C. spinosa var. spinosa was founded in fossils seeds from Iraq 5800 years before Christ. A worldwide study is necessary to confirm our hypothesis on Capparis evolution, speciation events and adaptation to geographic areas.
5 Conclusion
In conclusion, our study showed that AFLP analysis is an efficient method to investigate the genetic diversity and the relationship between species in Capparis. A total of six morphologically distinct species of caper were genotyped in Tunisia using 3 EcoRI–MseI AFLP primer combinations. Data analysis revealed a high degree of polymorphism allowing the distinction of the six species and their grouping into three main genetic clusters emphasizing the existence of recognizable genetic similarity within species and genetic heterogeneity between them. These three main clusters seem to be adapted to different geographic areas in correlation with bioclimate.
Acknowledgements
This study was carried out at Plant Biotechnology Laboratory of the Tunisian National Gene Bank. We are grateful to all reviewers for their very helpful comments on the manuscript.


