1 Introduction
Vinca or Catharanthus roseus is an important medicinal plant species with several applications in pharmaceuticals and industry. This plant species is well known for the biosynthesis of two anticancer bisindole alkaloids, namely vinblastine and vincristine, produced during the terpenoid indole alkaloids (TIAs) pathway [1,2]. This wild plant species can tolerate several biotic and abiotic stresses [3,4]; hence, we screened its transcriptome for usp-like gene family that might act in conferring tolerance to such adverse conditions. Although the exact function of this gene family is unknown, it acts not only through improving tolerance to biotic and abiotic stresses, but also tolerance to other stresses, e.g., starvation and DNA damage [5].
The RNA-Seq data of different tissues that were deposited in the CathaCyc database come from a study originally conducted by Van Moerkercke et al. [6]. This database was previously utilized [7] for detecting and characterizing 24 uspA-like genes. Transcription factors (TFs) with known actions that might drive these uspA-like genes likely indicate the mode of action of these genes. TFs are key proteins required in the regulation of almost all biosynthetic pathways in life [8]. They bind to specific target DNA sequences controlling cellular transcription by either promoting or blocking the recruitment of RNA polymerase to the target genes. TFs usually bind directly to the target promoter regions; however, they sometimes bind to RNA polymerase. The action of TF directly affect downstream genes functioning at a particular time on target cells (gene-in-time/gene-in-site). They are also important in driving the plant's responses to biotic and abiotic stresses [9].
In the present study, an in silico analysis was conducted in order to detect the TFs that might regulate the expression of uspA-like genes in C. roseus in a trial to detect the possible role of these uspA-like genes under biotic and abiotic stresses.
2 Materials and methods
An RNA-seq analysis of C. roseus available at NCBI (http://www.ncbi.nlm.nih.gov/sra, experiment SRP005953) was conducted in order to detect TFs concordantly expressed with uspA-like genes that had been previously characterized [7]. The data were de novo-assembled using Trinity-RNAseq v (r2013_08_14), and the recovered transcripts were calculated for abundance and differential expression using the Rsem package and the edgR method [7]. Blast alignment was done as previously described [7], and the values of fpkm for the recovered uspA-like transcripts and their co-expressed transcripts were calculated. The resulted transcripts were clustered based on their expression patterns across different tissues, treatments and hairy root genotypes. Clusters were studied in order to detect TF transcripts co-expressed with uspA-like transcripts in C. roseus. A structural analysis was done for selected C. roseus TF transcripts frequently co-expressed with uspA-like transcripts.
To validate the RNA-Seq dataset of C. roseus, semi-quantitative RT-PCR (or sqRT-PCR) was conducted for seven transcripts of selected clusters. They included four uspA-like genes and three co-expressing TFs. RNAs were isolated from four pot-grown plant tissues (e.g., mature leaf, flower, root and stem) using the Trizol reagent method (Invitrogen) and treated with RNase-free DNase (Promega Inc.). Primers were designed using Netprimer software (http://www.premierbiosoft.com/netprimer/index.html) with the following criteria: length of 20–27 bases, GC content of ∼50%, with minimal secondary structure, comparable annealing temperatures (48–55 °C) of the primer pairs, and PCR products of 281–300 bp (Table S1). sqRT-PCR was performed in forty cycles including denaturation at 94 °C for 15 s, annealing at 47–48 °C for 30 s, and extension at 72 °C for 45 s. Amounts of 1 ul cDNA, 1 × PCR buffer (with 1.5 mM MgCl2), 200 uM dNTPs, 200 nM of each gene-specific primers and 0.2 U of Taq DNA polymerase (Promega Inc.) were used in the reaction. Amplicons were run on a 1.2% agarose gels stained with ethidium bromide and visualized using the Gel Doc XR from Bio-Rad Laboratories (Hercules, CA, <). The actin gene (250 bp) was used as the house-keeping control.
3 Results and discussion
RNA-Seq analysis of the C. roseus SRA database of different tissues resulted in the recovery of 50,723 transcripts that were calculated for abundance and differential expression. A number of 24 uspA-like transcripts were previously detected and characterized [7]. The in silico analysis of the present study resulted in the recovery of 1245 transcripts involving 13, out of the 24, uspA-like transcripts (Table S2). The rest of the transcripts were not regulated in different tissues of C. roseus, hence not presented in the clusters generated from the analysis. The regulated transcripts were shown in 12 clusters based on their expression patterns across different tissues. BLAST to detect hits in the NCBI indicated the occurrence of a large number of TFs mainly in the erf (ethylene-responsive TF), bHLH (basic helix-loop-helix) and WRKY gene superfamilies. A number of 17 (four of erf, six of bHLH and seven of WRKY) TFs (see Table 1 for details) showed co-expression with six, out of the 13, uspA-like transcripts in five clusters, e.g., 4, 7, 8, 9 and 12 (Fig. 1 & Table S2). In cluster 4, the uspA-like transcript KJ634217 co-expressed with bHLH48. In cluster 7, the uspA-like transcripts KJ634222 and KJ634224 were in concordance with WRKY40, while the uspA-like transcript KJ634231 co-expressed with erf034. The uspA-like transcripts KJ634232 and KJ634221 co-expressed with a number of ten and five TFs of the three families, respectively. sqRT-PCR was used for validating the RNA-Seq dataset utilizing the selected co-expressing uspA-like and TF transcripts of clusters 4, 7 and 8 with varying expression levels in the four selected tissues, e.g., immature leaf (iML), mature leaf (ML), flower (FL), and root (R). The results concerning sqRT-PCR shown in Fig. 2 indicated an alignment with the RNA-Seq dataset (Table S2 and Fig. 1).
Transcription factors in Catharanthus roseus assemblies and their NCBI protein and mRNA hits.
| ID | Transcript ID | NCBI protein hit | Description |
| TF1 | comp30166_c0 | XP_004162335 | Cucumis sativus ethylene-responsive transcription factor erf034-like mRNA |
| TF2 | comp24014_c0 | XP_004164936 | Cucumis sativus ethylene-responsive transcription factor 5-like mRNA |
| TF3 | comp30976_c0 | XP_004173262 | Cucumis sativus ap2-like ethylene-responsive transcription factor bbm2-like mRNA |
| TF4 | comp16043_c0 | XP_004169980 | Cucumis sativus ethylene-responsive transcription factor erf008-like mRNA |
| TF5 | comp10162_c0 | XP_003521193 | Glycine max transcription factor bhlh48-like mRNA |
| TF6 | comp24246_c0 | XP_004250124 | Solanum lycopersicum transcription factor bhlh96-like mRNA |
| TF7 | comp15693_c0 | XP_006602463 | Glycine max transcription factor bhlh78-like transcript variant mRNA |
| TF8 | comp20416_c0 | XP_011460603 | Transcription factor bHLH106-like [Fragaria vesca subsp. vesca] |
| TF9 | comp2848_c0 | XP_006351643 | Solanum tuberosum transcription factor bhlh155-like transcript variant mRNA |
| TF10 | comp4341_c0 | XP_006356833 | Solanum tuberosum transcription factor bhlh74-like transcript variant mRNA |
| TF11 | comp10042_c0 | AJF11723 | Citrus sinensis probable wrky transcription factor 40-like mRNA |
| TF12 | comp11785_c0 | XP_003534796 | Glycine max probable wrky transcription factor 14-like transcript variant mRNA |
| TF13 | comp3515_c1 | XP_004235231 | Solanum lycopersicum probable wrky transcription factor 70-like mRNA |
| TF14 | comp20639_c0 | XP_004165456 | Cucumis sativus wrky transcription factor 72-like mRNA |
| TF15 | comp21556_c0 | XP_004504739 | Cicer arietinum wrky transcription factor 9-like transcript variant mRNA |
| TF16 | comp26488_c0 | XP_004501445 | Cicer arietinum wrky transcription factor 50-like mRNA |
| TF17 | comp8026_c1 | XP_004291261 | Fragaria vesca wrky transcription factor 11-like mRNA |
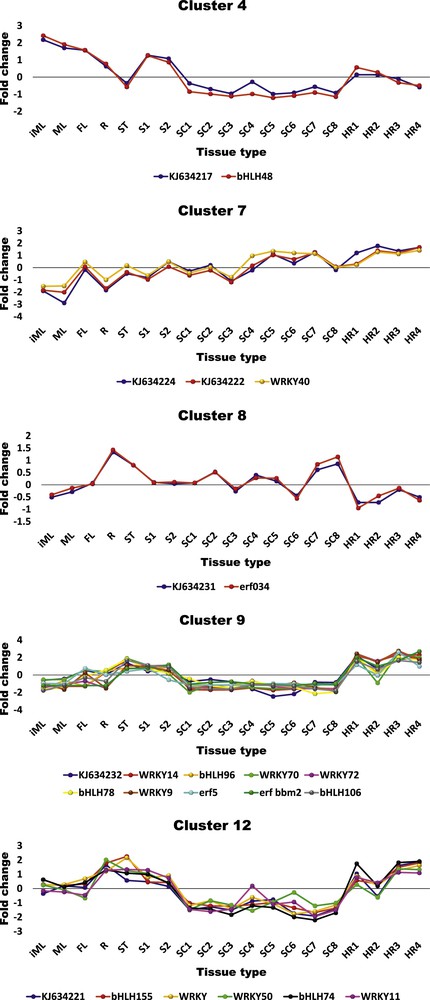
Co-expression of uspA-like transcripts and putative transcription factors (see Table 1 for details) of erf, bHLH and WRKY superfamilies in clusters 4, 7, 8, 9 and 12 (see Table S2) across Catharanthus roseus plant tissues. iML: immature leaf, ML: mature leaf, FL: flower, R: root, ST: stem, S: seedling, SC: suspension culture, HR: hairy root. TF1–4: transcription factors with erf domain, TF5–10: transcription factors with bHLH domain, TF11–17: transcription factors with WRKY domain.
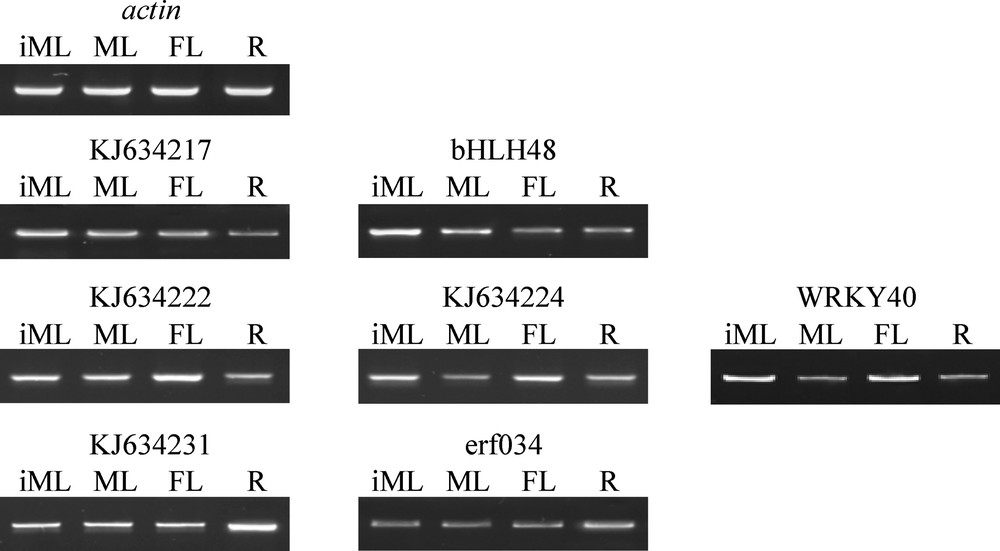
Semi-quantitative RT-PCR used for validating RNA-Seq analysis for three transcription factors (bHLH48, WRKY40 and erf034) and four uspA (IDs KJ634217, KJ634222, KJ634224 and KJ634231) genes of clusters 4, 7 and 8. Complete description of different TFs and co-expressing uspA-like genes and their expression patterns in immature leaf (iML), mature leaf (ML), flower (FL) and root (R) are shown in Table S2 and Fig. 1. The actin gene was used as the house-keeping control. Amplicons were run against 100-bp ladder DNA standard (Bioron). Masquer
Semi-quantitative RT-PCR used for validating RNA-Seq analysis for three transcription factors (bHLH48, WRKY40 and erf034) and four uspA (IDs KJ634217, KJ634222, KJ634224 and KJ634231) genes of clusters 4, 7 and 8. Complete description of different TFs and co-expressing Lire la suite
The AP-2 – or activating protein-2 of ethylene responsive (ER) TF – domain represents a superfamily of closely related TFs, which plays a critical role in regulating gene expression during the early development [10]. It was also shown to interfere with other signal transduction pathways [11,12]. Ethylene-responsive element-binding factors (ERFs) are a superfamily of transcriptional factors [13] that induce many developmental processes, and are also involved in the adaptation to biotic or abiotic stresses such as pathogen attack, wounding, UV irradiation, extreme temperature, and drought [14–16]. This TF is a key element in the integration of both signals for the regulation of defense response genes [17]. Recent studies have extended the influence of ethylene signaling with plant responses to both salt and water stress [18,19]. The erf8 and erf34 were reported in Arabidopsis and rice to be involved in the regulation of gene expression by stress factors and components of stress signal transduction pathways [13]. The erf was further reported to operate during the progression of leaf senescence [20], while erf5 was reported in Arabidopsis to regulate the chitin-induced innate immunity response [21]. Similar results were reached in tomato (Solanum lycopersicum), tobacco (Nicotiana tabacum), and Arabidopsis [22,23].
The information available for the basic helix-loop-helix (bHLH) superfamily indicates its existence in eukaryotes with numerous functions [24,25]. It can be responsible for the incoming regulatory signals from various cellular pathways and the direction of the transcriptional activity of a target gene [26]. The function of bHLH48 is not available, although this TF was characterized in a number of plant species including Arabidopsis (www.ncbi.nlm.nih.gov/gene/818831), rice (www.ncbi.nlm.nih.gov/gene/4341525), Glycine max (soybean) (www.ncbi.nlm.nih.gov/gene/100814124), corn (Zea mays) (www.maizegdb.org/gene_center/gene/bhlh48) and Jatropha (Jatropha curcas) (www.ncbi.nlm.nih.gov/gene/105634733). In Arabidopsis, bHLH74 was reported to regulate the expression of a subset of genes involved in cell expansion, while bHLH78 was reported to trigger flowering in response to blue light [27]. bHLH96 and bHLH155 have no known role, except that the expression of the first one was reported to be constitutive in roots, leaves, stems and flowers, while the second is involved in root development [24]. The bHLH106 was recently reported to participate in salt stress tolerance in Arabidopsis [28]. Knockout lines of this TF were more sensitive to NaCl, KCl, LiCl, ABA, and to low temperatures than the wild type.
The third type of TF, namely WRKY, is a member of a family of TFs functioning in plant responses to various physiological processes of vegetative and reproductive growth [29]. This type of TF is important in plant defense mechanisms, including MAMP-triggered immunity (MTI) or PAMP-triggered immunity (PTI), effector-triggered immunity (ETI), and systemic acquired resistance [30]. It was also proven to have a regulatory role in salt and mannitol stresses in Arabidopsis [31] and in cold and drought responses in barley (Hordeum vulgare) [32]. In addition, WRKY TFs regulate trichome and seed coat development in Arabidopsis [33], sesquiterpene biosynthesis in cotton (Gossypium hirsutum) [34], seed development in barley [35] and Arabidopsis [36], respectively, and senescence in Arabidopsis [37]. The WRKY1 of tobacco [38], similar to WRKY9 in Arabidopsis [39], was reported to be phosphorylated by the MAPK, namely SIPK, in order to mediate HR-like cell death. The WRKY11 and WRKY70 were reported more recently to participate in regulating the signal transduction pathways involved in triggering induced systemic resistance in Arabidopsis [40]. The WRKY14 was reported to act as a key regulatory role in methanol-induced tryptophan (Trp) and Trp-derived secondary metabolite biosynthesis during senescence in rice leaves [41]. The WRKY40 participates as transcriptional regulator in ABA signal transduction in Arabidopsis [42]. In the presence of the signal, WRKY40 relocalises to the nucleoplasm. Interestingly, the relocalisation was not observed in response to other abiotic or biotic stimuli. Recent reports indicated that WRKY50 is involved in low oleic acid- and SA-derived repression of jasmonic acid (JA)-inducible defense responses in Arabidopsis [43]. The WRKY72 was reported to participate in basal immunity in tomato and Arabidopsis, while appeared to be largely non-responsive to the defense hormone salicylic acid (SA) [44].
In general, we anticipate that the information in the present study highlights the possible functions of uspA-like genes in C. roseus in terms of cell response to biotic and abiotic stress. This is because a number of C. roseus uspA-like transcripts co-expressed with many putative ethylene-responsive, bHLH or WRKY putative TFs, whose analogues of some of them are known for their action in the plant defense against biotic and abiotic stresses.
Acknowledgments
This project was funded by the Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah, under Grant No. (15-3-1432/HiCi). The authors, therefore, acknowledge with thanks technical and financial support from DSR.


