1 Introduction
The marked toxicity of cadmium (Cd) ions on aquatic organisms with respect to the terrestrial counterparts is commonly attributed to their higher bioavailability in water [1] and to the multiple routes of uptake (skin, gills, gut) [2,3]. In both marine and freshwater habitats, Cd exerts its toxicity even at traces environmental concentrations, causing severe damage at the morphocytological level. Cd, for example, impairs tissue organization in the skeletal muscles [4,5], in the gills, liver and kidney [6–8], in the neuroglia in the central nervous system [9] and in the retina [10,11].
Cd exposure also alters gene expression [11–15] but the correlation existing between the observed molecular and morphological changes remains up to now rather unclear [16].
With the aim to identify early genes involved in cellular response to Cd stress in vertebrate embryos, we started a transcriptomic study using zebrafish as model organisms. The preliminary results herein described demonstrated a Cd-induction of the expression of the retinoblastoma binding protein 6 (RBBP6) and the crystallin lambda 1 protein (CRYL1) genes in gastrula embryos. RBBP6 protein is an E3 ubiquitin ligase involved in protein degradation [17]; the protein interacts with both p53 and pRb, promoting their degradation and thereby increasing cell proliferation. CRYL1 is a lens protein with enzymatic features first identified in rabbit and hare [18], down-regulated in hepatic cancer cells [19,20]. These two genes are both involved in eye development and in cellular response to cell diseases [21–23]. Since zebrafish eyes are particularly sensitive to Cd toxicity [10,11,24], we decided to investigate the expression of these 2 genes in zebrafish adult eyes by using in situ hybridization analysis, in natural conditions and after 30 days of Cd-exposure. In particular, an environmental concentration [5,11] and two sub-lethal concentrations (20 and 40 μM) were tested. To obtain information on the reversibility of the treatment, a group of animals exposed to 40 μM were maintained for 7 days in clean water and then subjected to in situ hybridization. The analyses demonstrated that both λ-crystallin and retinoblastoma binding protein 6 are Cd-sensitive and are upregulated by the metal in a dose-dependent manner.
Our data suggest an involvement of RBBP6 and CRYL1 proteins in the onset of morphological alterations previously observed in the zebrafish retina after exposure to cadmium.
2 Materials and methods
2.1 Animals and cadmium treatments
Adult and healthy zebrafish (Danio rerio) of both sexes were obtained from a specialized supplier and kept in standard conditions as previously described [25] in a 50-L housing tank for at least 2 weeks to acclimate before the experiments. Embryos were obtained as described by Westerfield [26]. Animal collection and housing were approved by the National Committee of the Italian ministry of health, and all experiments were conducted with the authority of the University Federico II ethical animal care and use committee. The experiments complied with the current laws of the European Union.
At the developmental stages of blastula (4 h after fertilization), an experimental group of 100 embryos were allowed to grow for 4 h in the embryo's medium added with CdCl2 for a final concentration of 20 μM Cd. Progression of embryonic development was scored by light microscopic observation. At the end of the treatment, embryos at the developmental stages of gastrula (approximately 80% epiboly) were washed, collected and processed for molecular analysis.
For the treatment of adult specimens, the animals were separated in four groups of 20 and housed in 10-L tanks with the respective treatment. The animals were transferred to the test aquarium filled with reverse osmosis water and kept in continuously aerated water, 25 ± 2 C, under a 14–10 h light/dark cycle photoperiod, fed three times a day with TetraMin Tropical Flake fish. The treatments were as follows: group 1, 1.5 μM Cd; group 2, 20 μM Cd; group 3, 40 μM Cd. The control group (group 4) was kept in the same conditions as the other groups, but without the addition of cadmium in the reverse osmosis water. Treatment was static (solutions remained unchanged throughout the duration of the test), lasted for 30 days and did not induce any evidence of physical or behavioral stress.
2.2 Total RNA isolation
Total RNAs from zebrafish embryos developed under natural (control) and Cd-contaminated (Cd-treated) conditions were extracted according to the TRI-REAGENT (Sigma Aldrich) protocol. Turbo DNA-free kit (Ambion) was used to digest the trace amounts of genomic DNA contamination in RNA. The two populations of total RNAs were dissolved in diethylpyrocarbonate (DEPC)-treated water and stored at −75 °C. The concentration and purity of RNA samples were determined by UV absorbance spectrophotometry; RNA integrity was checked using formaldehyde–agarose gel electrophoresis.
2.3 Differential display reverse transcriptase (RT)-PCR
DDRT-PCR was essentially performed as described previously [12,27]. Briefly, DNA-free total RNA (0.4 μg) extracted from either control or Cd-treated zebrafish embryos was reverse-transcribed in a 20-μL reaction mixture at 37 °C for 60 min with MMLV-reverse transcriptase and a set of two, one base anchored oligo(dT) primers. Amplification of cDNA fragments was performed using combinations of the anchored primers from the reverse-transcription step and 10 different upstream primers; all PCRs were repeated twice using the same cDNA sample. Aliquots (3 μL each) of amplification products were resolved on a 2% agarose gel (20 × 25 cm) with ethidium bromide in TAE buffer. Using a sharp, clean razor blade, a rectangular piece of gel corresponding to an individual band of interest was excised and the cDNA fragment recovered using a gel extraction kit (5PRIME). The eluted cDNA was re-amplified in a PCR reaction using the same pair of primers used in the differential display reaction. The Cd-responsive fragments were inserted into a pCRII-TOPO vector (Invitrogen) and cloned following the manufacturer's instructions. Recombinant plasmid DNA was sequenced bidirectionally by the DNA sequencing service of Primm Biotech, using T7 and T3 primers. The homology search of genes was performed by online-based FASTA and BLAST programs through the European Molecular Biology Laboratory (EMBL) nucleotide sequence database at European Bioinformatics Institute (http://www.ebi.ac.uk).
2.4 In situ hybridization
At the end of the treatments, adult zebrafish were anesthetized with MS222 (tricaine methanesulfonate, 1:15,000 w/v) and sacrificed by decapitation. Heads were rapidly dissected, fixed in Bouin's solution and processed for paraffin wax embedding according to routine protocols [28]. Sections (5–7 μm) were mounted on superfrost glass slides (Menzel-Glaser, Germany) and used for in situ hybridization [29]. In particular, they were fixed in paraformaldehyde 4% PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4) pH 7.4 for 20 min, and incubated in PK buffer (Tris-HCl, 0.2 M, pH 7.4, EDTA 0.01 M, pH 8, proteinase K, 10 mg/mL, H2Odepc) at 37 °C for 15 min. After washing in PBS, they were incubated at 42 °C for 90 min in a prehybridization mix containing formamide, SSC 4× and 1× Denhart's solution. Hybridization was carried out at 42 °C overnight using a dig-labeled cDNA probe encoding a fragment of Danio rerio RBBP6 or CRYL1. Sections were washed in SSC 2× (Tris-HCl 0.1 M, pH 7.5, NaCl 0.1 M, H2Odepc) and in Buffer I containing the blocking reagent (0.5%). Digoxigenin was revealed by incubating sections with an AP-conjugated anti-dig antibody diluted 1:400 overnight. Slides were washed in Buffer I, incubated with levamisole-Tween20 1× for 15 min and revealed with BM-Purple. Dig-labeled cDNA probes were generated by PCR using the DIG High Prime DNA labeling and detection starter kit I (Roche Diagnostics, Germany). In the negative control, the hybridization solution did not contain the cDNA probe.
3 Results
3.1 Identification of CRYL1 and RBBP6 as Cd-responsive genes in zebrafish embryos
Comparison of mRNA expression patterns between control and Cd-treated zebrafish gastrulae allowed us to identify two cDNA fragments whose expression were upregulated following Cd-exposure (Fig. 1).
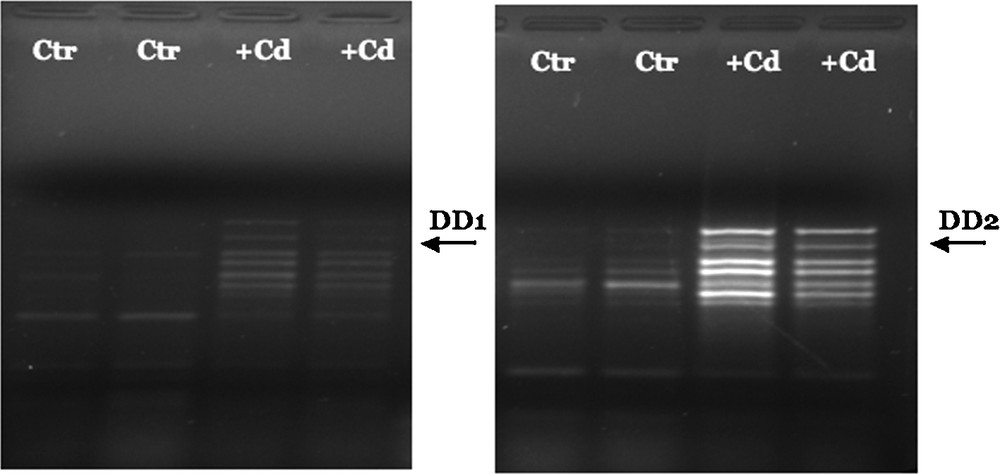
Representative differential display electrophoresis of mRNA from control and Cd-treated zebrafish. Total RNA from control and Cd-treated embryos was reverse-transcribed using the 3’-primer oligo (dT) H-T11A: 5’-AAGCTTTTTTTTTTTA. DD1 fragment was amplified by PCR reaction using the H-T11A primer and the 5’-random primer: 5’-AAGCTTCTCAACG-3’; DD2 fragment using the H-T11A primer and the 5’-random primer: 5’-AAGCTTTGGTCAG-3’. PCR reactions were performed in duplicate for each cDNA sample to minimize artefacts. The arrows indicate the two cDNA bands that were recovered from the gels and analyzed further.
The two fragments were re-amplified, cloned and sequenced; the retrieved nucleotide sequences were compared against EMBL nucleotide sequence database. The results of BLAST analysis, reported in Table 1, clearly showed that the clone DD1 corresponds to a 254 bp fragment of the D. rerio gene encoding crystallin lambda 1 (CRYL1) and the clone DD2 corresponds to a 161 bp fragment of the D. rerio gene for the retinoblastoma protein binding protein 6 (RBBP6).
Blast analysis of Cd-regulated cDNAs.
| Clone | Base pairs | Effect of cadmium | Sequence homology | EMBL accession number | Identity (%) | E-score |
| DD1 | 254 | + | Danio rerio crystallin, lambda 1, mRNA | BC121722 | 99.6 | 6.9−124 |
| DD2 | 161 | + | Danio rerio retinoblastoma protein binding protein 6, mRNA | BG737479 | 99.2 | 6.6−64 |
3.2 Effects of cadmium on the expression of CRYL1 gene in zebrafish adult eyes
Cadmium treatment did not induce significant morphological alterations; treated retinae in fact, showed only occasional and modest disorganized areas. These extended for a few square microns, and were more usually located in the outer layers, involving in particular the pigmented epithelium that appeared dislocated into the nervous retina (Fig. 2C, G).
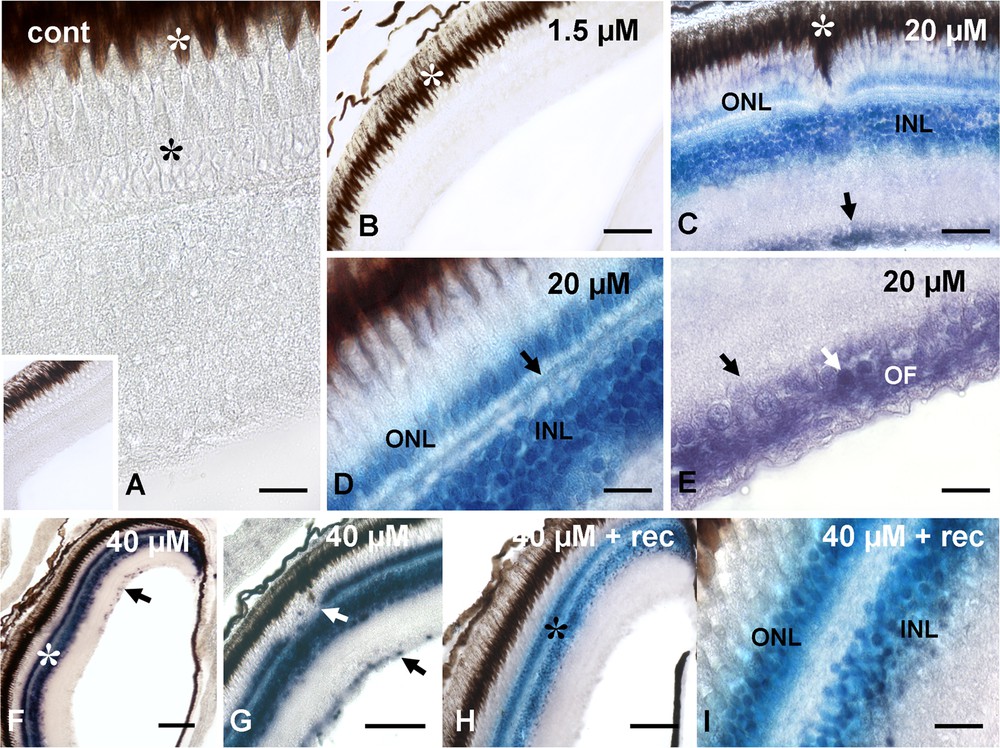
Localization of CRYL1-mRNA in retinae of control (A) and cadmium treated (B-I) Danio rerio. A and inset and (B) unlabeled retinae. Notice the photoreceptors (*) and the retinal pigmented epithelium (white *). (C) Labeling of inner (INL) and outer (ONL) nuclear layers and of ganglion cell layer (arrow). Note the partially dislocated pigmented epithelium (white *). (D) Detail of labeled nuclear layers and horizontal cells (arrow). (E) Ganglion cell layer; labeled nuclei (white arrow) and optic fiber (OF) surrounding unlabeled nuclei (arrow). (F) Labeled nuclear layers (*) and occasional ganglion cell nuclei (arrow). (G) Detail of labeled nuclear layers. Notice the interrupted outer nuclear layer (white arrow) and the labeled ganglion cell nuclei (arrow). (H) Labeled nuclear layers (*). (I) Detail. Bars: A, D, e, I: 10 μM; C: 25 μM; B, F–H: 50 μM.
After in situ hybridization, all control (Fig. 2A) and 1.5-μM-cadmium-treated (Fig. 2B) retinae were consistently negative. In contrast, the retinae of animals exposed to cadmium at 20 μM showed a significant labeling (Fig. 2C), localized on the inner and outer nuclear layers (Fig. 2C), on the optical fibers and on the scattered ganglion cell nuclei (Fig. 2E). At higher magnification (Fig. 2D), labeling was also observed on the horizontal cells of the outer plexiform layer.
The retinae of animals exposed to cadmium at 40 μM had labeled inner and outer nuclear layers (Fig. 2F) and, to a lesser extent, optic fibers (Fig. 2G). The nuclei in the ganglionic layer were only occasionally labeled (Fig. 2F and G), and the horizontal cells were always unlabeled as most of the optic fibers (Fig. 2G).
Exposure of these animals to a 7-day period of recovery in uncontaminated water induced no substantial changes in labeling localization. The two nuclear layers and the horizontal cells (Fig. 2G–I) remained labeled and unlabeled, respectively. In contrast, in the ganglionic and optical layers, labeling completely disappeared (Fig. 2H).
3.3 Effects of cadmium on the expression of RBBP6 genes in zebrafish adult eyes
Control (Fig. 3A) and 1.5 μM cadmium treated retinae (Fig. 3B) were always unlabeled in contrast to retinae of animals treated with 20 μM Cd that had labeled inner nuclear layers. Labeling distribution and intensity were clearly non-uniform, being more intense close to the emergence of the optic nerve (Fig. 3C) and in the distal retina (Fig. 3D and E). The outer nuclear layer, the plexiform layers and the ganglionic cell layer were always unstained (Fig. 3C–F). The labeling distribution did not change in correspondence of morphologically altered areas (Fig. 3F).
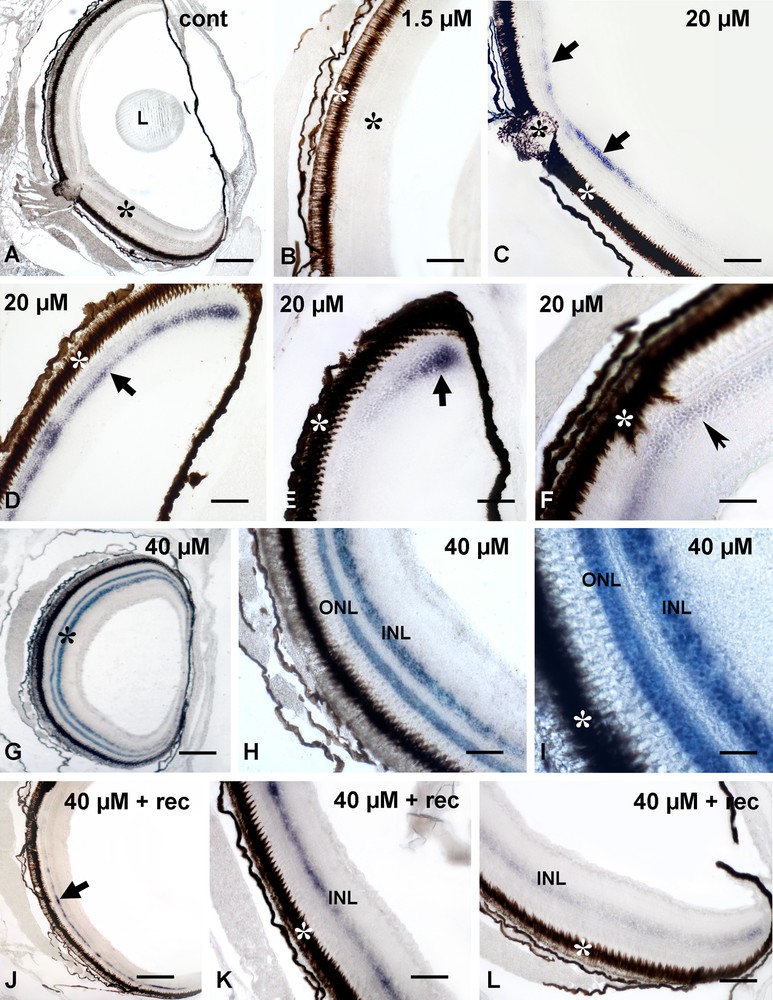
Localization of RBBP6-mRNA in retinae of control (A) and cadmium treated (B–L) Danio rerio. (A) and (B) unlabeled retinae (*); lens (L) and retinal pigmented epithelium (white *); (C) moderate and discontinuous labeling on the inner nuclear layer (arrows). Retinal pigmented epithelium (white *) and emergence of the optic nerve (*). (D) Distal retina with labeled inner nuclear layer (arrow). (E) Detail of a distal anterior retina with a spot of intensely labeled cells (arrow). (F) Dislocated pigmented epithelium (white *) and faintly labeled inner nuclear layer (*). (G) Intensely labeled nuclear layers (*). (H) Detail showing the outer (ONL) and inner (INL) nuclear layers. (I) Further detail. (J) Faintly and discontinuously labeled inner nuclear layer (arrow). Detail of labeled inner nuclear layer (INL) in the proximal (K) and distal (L) retina. Bars: A, G, J: 120 μM; E–F, H–I, 25 μM; B–D, K–L: 50 μM.
The retinae of 40-μM-Cd-treated animals showed an intense labeling distributed consistently on both inner and outer nuclear layers (Fig. 3G–I). Plexiform and ganglionic cell layers remained always unstained (Fig. 3H).
Exposure of 40 μM Cd treated animals to a 7-day period of recovery in uncontaminated water induced a significant decrease in labeling (Fig. 3J–L). The outer nuclear layer was completely unstained, while the inner nuclear layer appeared faintly and discontinuously labeled, with a pattern very much resembling that observed in 20 μM Cd treated retinae.
4 Discussion
It is widely demonstrated that cadmium alters/deregulates gene expression in animal tissues [12,27,30]; nevertheless, the exact mechanism and the consequences of the changes induced are not always clear. In some cases, even the nature of the proteins/genes up/down-regulated is unknown [12,27].
The data herein described contribute to the comprehension of the molecular response to cadmium intoxication by identifying another two genes, CRYL1 and RBBP6, that in Danio rerio are upregulated by metal intoxication in a dose- and time-dependent manner. This occurs at the gastrula stage and, in the adult, in the retina, a target organ for cadmium toxicity [10,11].
RBBP6 is a protein primarily involved in the control of apoptosis and cell differentiation [17,31], and, therefore, it is not surprising that it is over-expressed in the two nuclear cell layers. In Danio, the photoreceptors and the inner nuclear layer cells are extensively damaged by cadmium exposure, eliminated via apoptosis and substituted by newly differentiated glial Muller cells [11]. RBBP6-driven cells replacement would contribute to the restoration of the retinal integrity, avoiding more significant structural alterations comparable to those occurring, for example, in lizard embryos, where extensive folding is reported [32] or in invertebrates, such as the crustacean nauplii [33,34].
The same apparently does not occur in ganglion cells in which cytoplasm vacuolization and nuclear pycnosis have also been reported [11], but the expression of RBBP6 is not activated. A possible explanation is in the origin of the different retinal components. In the embryo, a first wave of neurogenesis originates the ganglion cells, while other proneural genes will lead to the differentiation of inner and outer nuclear cells during another two successive differentiation waves [35]. Being this considered, it appears possible that ganglion cells have a different, genetic, susceptibility to metal contamination. No data are available, as far as we are aware, about the transcriptional activity of RBBP6 gene or about the presence of the protein, in this retinal layer in normal eyes.
Lambda-crystallin 1 has a completely different function with respect to RBBP6 being involved in NADH-binding [36] and in the control of the redox state [37]. For these reasons, it is constitutively expressed in the retinal nuclear and ganglion cells layers of vertebrates [38], where it would protect from light damage [39]. In cadmium-exposed Danio, the oxidative stress is high [40] and it is conceivable that increased crystallin levels will match redox homeostasis needs. Overexpression of lambda-crystallin 1, in effect, is induced in several models, including isolated clonal cells lines obtained from tumors, indicating that the gene is part of a distinctive expression profile induced by the ion [41].
What is unexpected, in Danio, is the absence of messengers in controls. Low levels of transcript and/or a rapid messenger turnover may be postulated to explain the observation, but further investigations aiming at determining the protein concentration are needed to clarify such a striking difference with other species [38].
In conclusion, it can be postulated that the upregulation of RBBP6 and CRYL-1 induced by cadmium exposure helps in protecting the retina, and maybe also the embryos, of Danio rerio from metal cytotoxicity.
Disclosure of interest
The authors declare that they have no competing interest.


