1 Introduction
Molecular markers, such as inter simple sequence repeat (ISSR), random amplified polymorphic DNA (RAPD), simple sequence repeats (SSR) and amplified fragment length polymorphism (AFLP) are routinely used in ecological, evolutionary, taxonomical, phylogenic and genetic studies in plant science [1]. ISSR is a technique that involves the use of microsatellite sequences as primers in a polymerase chain reaction (PCR) to generate multilocus markers [2]. RAPD is a technique based on the amplification of discrete regions of the genome by PCR with short oligonucleotide primers of arbitrary sequence [3]. As these two approaches require no prior knowledge of the genome that is being analyzed, they can be used across species using universal primers. Due to their speed and efficiency, both are simple methods that combine many of the advantages of SSR and AFLP [4]. ISSR and RAPD have been successfully used to estimate the extent of genetic diversity at inter- and intra-specific levels in a wide range of plant species [5–7]. As ISSR and RAPD are PCR-based techniques, factors such as the concentration of dNTPs and TaqDNA polymerase, annealing temperature, extension time, cycle times and the integrity of the template may influence the PCR and affect ISSR and RAPD analysis. Although an optimization of PCR experiment reagents and methods can improve these analyses, the integrity of the template is determined by the method of preservation prior to DNA isolation. The preservation of plant materials is a major obstacle in ISSR and RAPD analyses, as plants are often collected far from laboratory facilities.
Preservation techniques for plant tissues incorporate three main categories: chemical treatments, cryopreservation, and dried preservation. Chemical treatments, such as formalin-acetic acid-ethanol (FAA), ethanol, Carnoy's solution, and chloroform ethanol, are suitable for fixing plant tissues, but cause DNA degradation, since several of these preservatives contain acids or other denaturants that can destroy DNA integrity [8,9]. Cryopreservation is the storage of viable cells, tissues, organs and organisms at ultra-low temperatures, usually in liquid nitrogen to a minimum temperature of −196 °C [10], and is widely used for the long-term storage of plant germplasm [11–14]. Drying specimens in the field has proven successful for subsequent extraction and can be done by air drying [15] or rapid drying of plant material in silica gel. However, air drying gives poor results for several species in the Malpighiaceae and Orchidacea, for which DNA was obtained but highly degraded [16]. Preservation in silica gel is most appropriate as a general drying method [9,16]. Approximately 50 g of silica gel will reliably desiccate 1 g fresh weight of leaf material for most species of higher plants [17].
We used the preservation methods of drying in silica gel, preservation in liquid nitrogen and preservation in a −20 °C ice chest when investigating the genetic structure of Rhododendron aureum Georgi (Ericaceae) [18] and Rhodiola sachalinensis (endemic) [19]. In these two studies, we found that different preservation methods on the same sample resulted in different ISSR and RAPD loci. Thus, the method of drying in silica gel was selected for these studies to eliminate these effects. ISSR and RAPD analysis may be affected by different preservation methods for other plant materials. In this paper, we address this issue by comparing three frequently-used preservation methods for plant materials in six species of three of the most represented families of broad-leaved, needle-leaved and succulent plants for ISSR and RAPD analysis. Furthermore, we compare the effects between samples preserved for three weeks and three years in an effort to find a method suitable for plant material characteristics and provide a reference for researchers who wish to analyze variation at the DNA level as part of their research.
2 Materials and methods
2.1 Sampling
We sampled six species, the broad-leaved trees Populous tomentosa Carr. (Y) and Salix babylonica L. (L), the needle-leaved trees Pinus koraiensis Sieb. (H) and Pinus sylvestris var. mongolica Litv. (S), and the succulent herb Hylotelephium spectabile (Bor.) H. Ohba (T) and Sedum sarmentosum Bunge (C), from the campus of Jilin University between June and July 2013. These leaves were at a similar growing status and free from fungus, other infections or dirt. We used three preservation methods to preserve the samples; they were divided into three groups and stored in cryogenic liquid nitrogen (N2), preserved in a −20 °C ice chest (I), and rapidly dried in silica gel (D). In order to rule out the influence of individual genetic difference and experimental bias, each species was derived from the same individual, and 16 specimens were sampled. Eight samples from each group were preserved for three weeks, while the other 8 samples were preserved for three years.
2.2 DNA extraction and ISSR/RAPD-PCR
We isolated genomic DNA of all the preserved samples and extracted fresh leaves for use as the control using the plant genomic DNA kit (Tiangen Biotech. Beijing Co. Ltd., China), following the manufacturer's instructions. Each preservation method for all species was repeated 8 times in case of random errors. DNA quality and quantity were estimated both spectrophotometrically, as well as visually, by ethidium bromide staining after electrophoresis on 0.8% agarose gels. The DNA samples were diluted to the concentration of 50 ng/μL and stored at −20 °C for use. A set of 30 anchored microsatellite primers and 20 decamer arbitrary primers were synthesized from Sangon Biotech. Beijing Co. Ltd., China, and at least 8 primers were selected for each species (see Tables S1–S4). ISSR–PCR was performed in 25-μL PCR reaction tubes containing 2 μL of genomic DNA, 1.0 μL of primer (20 μM stock, 0.8 μM final concentration), 3.0 μL of dNTP mix (2.5 mM stock, 0.3 mM final concentration), 2.5 μL of standard Taq reaction buffer (10×), and 0.2 μL of Taq DNA polymerase enzyme (5000 unit/mL; TransGen Biotech, China). Thermocycling proceeded as follows: 94 °C for 5 min followed by 40 cycles at 94 °C for 45 s, 49–59.8 °C annealing for 45 s, 72 °C for 1 min; completion of these cycles was followed by a final extension at 72 °C for 10 min. The annealing temperature was usually adjusted according to the Tm of the primer used in the reaction. RAPD-PCR was performed in a 25-μL volume having 1× PCR buffer, dNTP (0.2 mM), primer (10 pmol), 5 ng template and Taq DNA polymerase (1 U) (TransGen Biotech, China). RAPD amplification was performed with initial denaturation at 94 °C for 5 min, followed by 45 cycles of 1 min denaturation at 94 °C, 1 min annealing at 37 °C, 1 min extension at 72 °C and final extension at 72 °C for 10 min. The amplified products were mixed with bromophenol blue gel loading dye and were analyzed by electrophoresis on a 1.2% agarose gel using a 0.5× Tris Acetate EDTA buffer pH 8.0 at room temperature. In general, the quality of the patterns generated differed from primer to primer (Fig. S1).
2.3 Data analysis
The amplified fragments, with the same mobility according to their molecular weight (bp), were scored according to whether a binary code was present (1) or absent (0). Only the consistently reproducible bands were scored, and smeared and weak bands were excluded. The number of bands from each group was manually counted and least significant difference (LSD) multiple comparisons were performed with SPSS (version 19). In order to detect the influence of different preservation methods for plant materials on ISSR and RAPD analysis, we calculated the genetic index used for each molecular marker. The statistical analysis of ISSR and RAPD markers were analyzed using POPGENE software (version 1.31) [20]. An analysis of molecular variance (AMOVA) was used to examine population genetic differentiation within and among the seven groups preserved by different methods during different times using AMOVAPREP and AMOVA 1.55 [21]. The dendrograms were constructed using an unweighted paired group method of cluster analysis with arithmetic averages (UPGMA) on NTSYS-pc program (version 2.10e) [22].
3 Results
3.1 The number of bands detected by ISSR and RAPD markers
All the materials preserved in the seven groups resulted in sufficient levels of genomic DNA for use in ISSR and RAPD analysis. However, there were some differences in the number of bands (Figs. 1 and 2), except for P. koraiensis analyzed by RAPD, which resulted in 67 bands in all groups (Fig. 2c). In the ISSR analysis, the fresh samples of salicaceae and pinaceae showed the more bands. In particular, there were significantly more bands in the fresh pinaceae group than in other groups. Crassulaceae preserved in liquid nitrogen showed the more bands; the range of bands between the highest and lowest groups was higher than that in salicaceae and pinaceae. Significant differences were observed between short-time and long-time preservation, storage in liquid nitrogen of Se. sarmentosum and storage in −20 °C ice chest of Po. tomentosa, S. babylonica, and P. sylvestris var. mongolica. However, no significant differences were evident using other methods. In RAPD analysis, fresh samples of Po. tomentosa showed the more bands, but the number of bands was not significantly different from that of samples preserved in liquid nitrogen (Fig. 2a); S. babylonica preserved in liquid nitrogen and dried in silica showed more bands; P. sylvestris var mongolica preserved in liquid nitrogen showed significantly more bands than other groups; with regards to crassulaceae, the fresh samples showed the least number of bands and no significant difference between other H. spectabile samples was observed. The results show that different preservation methods have wide-ranging effects on the number of bands of different plant types, in both ISSR and RAPD analyses.
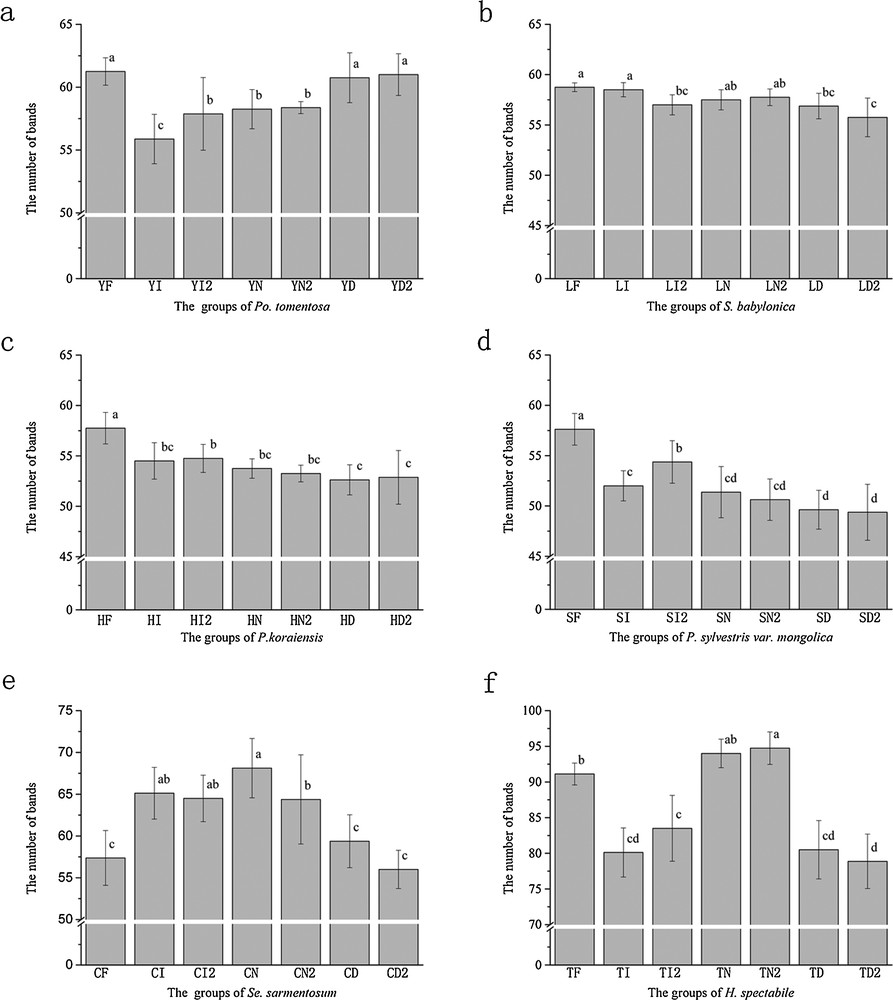
Number of bends detected by ISSR analysis. There were significant differences among the groups with the different letters (<0.05), and there were no significant differences among the groups with the same letter. Bars are standard errors.
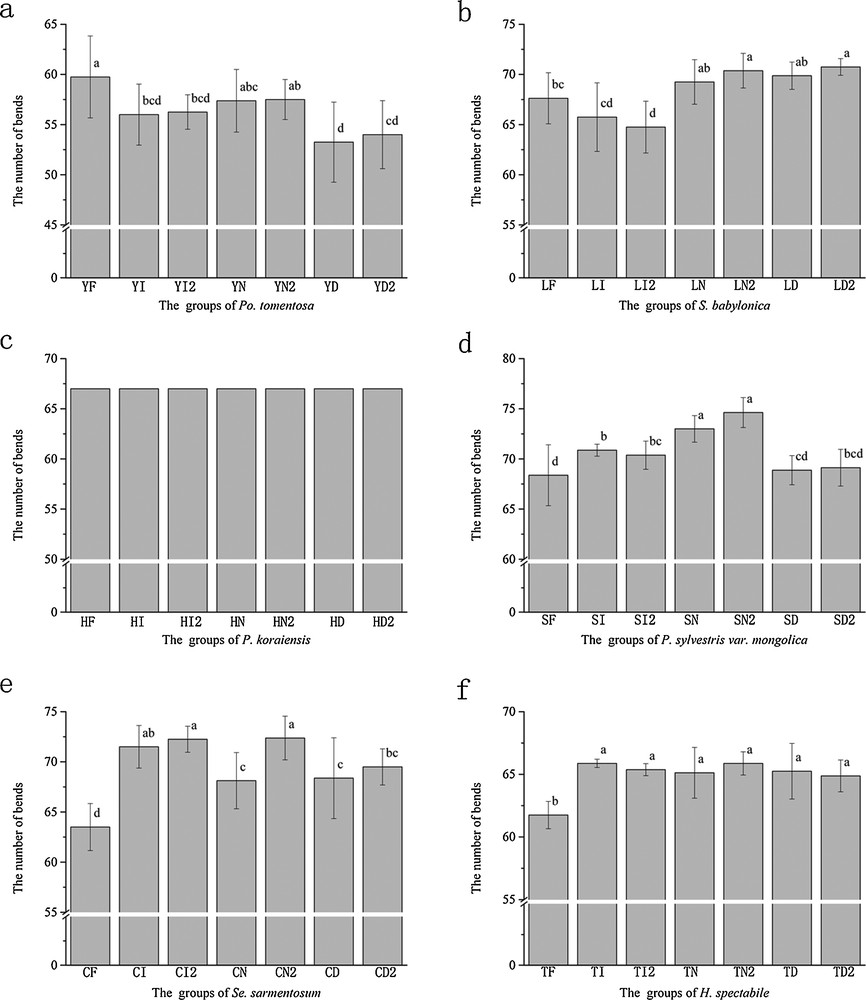
Number of bends detected by RAPD analysis. There were significant differences among the groups with the different letters (<0.05), and there were no significant differences among the groups with the same letter. Bars are standard errors.
3.2 Genetic diversity index by ISSR and RAPD markers
The genetic diversity detected by ISSR and RAPD is shown in Table 1. In the ISSR analysis, Po. tomentosa and H. spectabile, preserved in a −20 °C ice chest, showed the highest percentage of polymorphic loci (PPL) and the highest Shannon information index (i), whereas S. babylonica, P. koraiensis, and Se. sarmentosum, which were preserved by drying in silica gel, and P. sylvestris var mongolica, which were preserved in liquid nitrogen and dried in silica gel, showed a higher PPL. In the RAPD analysis, Po. tomentosa, which was preserved in liquid nitrogen, showed the most PPL and for S. babylonica was preserved in −20 °C ice chest; the fresh and dried in silica groups of P. koraiensis and H. spectabile got the higher PPL; the fresh groups of Se. sarmentosum, preserved in liquid nitrogen got higher PPL values; the groups of P. sylvestris var mongolica preserved in liquid nitrogen groups and dried in silica got higher PPL values.
Genetic diversity detected by ISSR and RAPD analysis of the selected six species preserved by three methods for different time.
| Group | ISSR | RAPD | ||||||||||
| n | PPL | n a | n e | h | i | n | PPL | n a | n e | h | i | |
| YF | 12 | 14.12 | 1.14 | 1.08 | 0.05 | 0.07 | 26 | 32.10 | 1.32 | 1.25 | 0.14 | 0.19 |
| YI | 19 | 22.35 | 1.22 | 1.15 | 0.08 | 0.12 | 25 | 30.86 | 1.31 | 1.19 | 0.11 | 0.16 |
| YI2 | 18 | 21.18 | 1.21 | 1.14 | 0.08 | 0.12 | 21 | 25.93 | 1.26 | 1.16 | 0.09 | 0.13 |
| YN | 10 | 11.76 | 1.12 | 1.08 | 0.04 | 0.06 | 28 | 34.57 | 1.35 | 1.25 | 0.14 | 0.20 |
| YN2 | 7 | 8.24 | 1.08 | 1.04 | 0.02 | 0.04 | 26 | 32.10 | 1.32 | 1.20 | 0.12 | 0.17 |
| YD | 13 | 15.29 | 1.15 | 1.09 | 0.05 | 0.08 | 25 | 30.86 | 1.31 | 1.15 | 0.09 | 0.14 |
| YD2 | 12 | 14.12 | 1.14 | 1.09 | 0.05 | 0.08 | 23 | 28.40 | 1.28 | 1.14 | 0.09 | 0.14 |
| LF | 3 | 4.48 | 1.04 | 1.03 | 0.02 | 0.03 | 18 | 21.95 | 1.22 | 1.17 | 0.09 | 0.13 |
| LI | 3 | 4.48 | 1.04 | 1.04 | 0.02 | 0.03 | 24 | 29.27 | 1.29 | 1.23 | 0.13 | 0.18 |
| LI2 | 8 | 11.94 | 1.12 | 1.09 | 0.05 | 0.07 | 20 | 24.39 | 1.24 | 1.17 | 0.09 | 0.14 |
| LN | 9 | 13.43 | 1.13 | 1.07 | 0.04 | 0.07 | 19 | 23.17 | 1.23 | 1.17 | 0.09 | 0.14 |
| LN2 | 8 | 11.94 | 1.12 | 1.07 | 0.04 | 0.06 | 14 | 17.07 | 1.17 | 1.10 | 0.06 | 0.09 |
| LD | 12 | 17.91 | 1.18 | 1.09 | 0.06 | 0.09 | 16 | 19.51 | 1.20 | 1.15 | 0.08 | 0.12 |
| LD2 | 14 | 20.90 | 1.21 | 1.13 | 0.08 | 0.11 | 8 | 9.76 | 1.10 | 1.07 | 0.04 | 0.06 |
| HF | 19 | 22.62 | 1.23 | 1.11 | 0.07 | 0.11 | 11 | 15.07 | 1.15 | 1.13 | 0.07 | 0.09 |
| HI | 12 | 14.29 | 1.14 | 1.08 | 0.05 | 0.07 | 4 | 5.48 | 1.05 | 1.03 | 0.02 | 0.03 |
| HI2 | 10 | 11.90 | 1.12 | 1.08 | 0.04 | 0.07 | 5 | 6.85 | 1.07 | 1.04 | 0.02 | 0.04 |
| HN | 18 | 21.43 | 1.21 | 1.15 | 0.08 | 0.12 | 6 | 8.22 | 1.08 | 1.06 | 0.03 | 0.05 |
| HN2 | 19 | 22.62 | 1.23 | 1.14 | 0.08 | 0.12 | 10 | 13.70 | 1.14 | 1.10 | 0.06 | 0.08 |
| HD | 21 | 25.00 | 1.25 | 1.16 | 0.09 | 0.13 | 9 | 12.33 | 1.12 | 1.11 | 0.06 | 0.08 |
| HD2 | 31 | 36.90 | 1.37 | 1.19 | 0.11 | 0.17 | 11 | 15.07 | 1.15 | 1.13 | 0.07 | 0.10 |
| SF | 24 | 28.57 | 1.29 | 1.19 | 0.11 | 0.16 | 23 | 27.06 | 1.27 | 1.22 | 0.12 | 0.17 |
| SI | 20 | 23.81 | 1.24 | 1.15 | 0.09 | 0.13 | 3 | 3.53 | 1.04 | 1.02 | 0.01 | 0.02 |
| SI2 | 22 | 26.19 | 1.26 | 1.17 | 0.10 | 0.14 | 7 | 8.24 | 1.08 | 1.06 | 0.03 | 0.05 |
| SN | 33 | 39.29 | 1.39 | 1.25 | 0.15 | 0.22 | 11 | 12.94 | 1.13 | 1.09 | 0.05 | 0.08 |
| SN2 | 24 | 28.57 | 1.29 | 1.17 | 0.10 | 0.15 | 9 | 10.59 | 1.11 | 1.09 | 0.05 | 0.07 |
| SD | 30 | 35.71 | 1.36 | 1.21 | 0.13 | 0.19 | 11 | 12.94 | 1.13 | 1.10 | 0.06 | 0.08 |
| SD2 | 31 | 36.90 | 1.37 | 1.22 | 0.13 | 0.19 | 11 | 12.94 | 1.13 | 1.10 | 0.06 | 0.08 |
| CF | 28 | 29.17 | 1.29 | 1.17 | 0.10 | 0.15 | 28 | 29.17 | 1.29 | 1.18 | 0.10 | 0.15 |
| CI | 29 | 30.21 | 1.30 | 1.17 | 0.10 | 0.16 | 10 | 10.42 | 1.10 | 1.07 | 0.04 | 0.06 |
| CI2 | 27 | 28.12 | 1.28 | 1.16 | 0.09 | 0.14 | 10 | 10.42 | 1.10 | 1.06 | 0.04 | 0.05 |
| CN | 28 | 29.17 | 1.29 | 1.21 | 0.12 | 0.17 | 28 | 29.17 | 1.29 | 1.17 | 0.10 | 0.15 |
| CN2 | 35 | 36.46 | 1.36 | 1.25 | 0.14 | 0.21 | 22 | 22.92 | 1.23 | 1.18 | 0.10 | 0.14 |
| CD | 27 | 28.12 | 1.28 | 1.20 | 0.11 | 0.16 | 24 | 25.00 | 1.25 | 1.19 | 0.10 | 0.15 |
| CD2 | 38 | 39.58 | 1.40 | 1.27 | 0.15 | 0.22 | 22 | 22.92 | 1.23 | 1.15 | 0.09 | 0.13 |
| TF | 26 | 18.44 | 1.18 | 1.12 | 0.07 | 0.10 | 8 | 11.43 | 1.11 | 1.07 | 0.04 | 0.06 |
| TI | 45 | 31.91 | 1.32 | 1.21 | 0.12 | 0.17 | 1 | 1.43 | 1.01 | 1.01 | 0.01 | 0.01 |
| TI2 | 50 | 35.46 | 1.35 | 1.24 | 0.14 | 0.20 | 2 | 2.86 | 1.03 | 1.03 | 0.01 | 0.02 |
| TN | 29 | 20.57 | 1.21 | 1.16 | 0.09 | 0.13 | 7 | 10.00 | 1.10 | 1.08 | 0.04 | 0.06 |
| TN2 | 25 | 17.73 | 1.18 | 1.13 | 0.07 | 0.11 | 6 | 8.57 | 1.09 | 1.07 | 0.04 | 0.05 |
| TD | 42 | 29.79 | 1.30 | 1.18 | 0.10 | 0.15 | 8 | 11.43 | 1.11 | 1.09 | 0.05 | 0.07 |
| TD2 | 39 | 27.66 | 1.28 | 1.15 | 0.09 | 0.14 | 7 | 10.00 | 1.10 | 1.07 | 0.04 | 0.06 |
3.3 AMOVA for ISSR and RAPD data
The genetic variation attributed among the groups deviated from 50%, in particular P. sylvestris var mongolica, Se. sarmentosum, and S. babylonica with both ISSR and RAPD markers (Table 2). Only H. spectabile samples had variation of nearly 50% among the groups. However, the similar genetic variation between groups and within groups does not indicate that the different methods had no effect on ISSR or RAPD analysis.
Analysis of molecular variance (AMOVA) for ISSR and RAPD data from selected six species.
| Species | Source of variation | d.f. | ISSR | RAPD | ||||
| SSD | Variance components | Variation (%) | SSD | Variance components | Variation (%) | |||
| Y | a.g. | 6 | 171.54 | 3.25 | 55.98 | 208.00 | 3.72 | 43.16 |
| w.g. | 49 | 125.38 | 2.56 | 44.02 | 240.13 | 4.90 | 56.84 | |
| L | a.g. | 6 | 63.61 | 1.12 | 41.21 | 119.82 | 2.07 | 38.02 |
| w.g. | 49 | 78.63 | 1.60 | 58.79 | 165.63 | 3.38 | 61.98 | |
| H | a.g. | 6 | 228.50 | 4.31 | 54.17 | 106.96 | 2.05 | 59.40 |
| w.g. | 49 | 178.50 | 3.64 | 45.83 | 68.75 | 1.40 | 40.60 | |
| S | a.g. | 6 | 201.32 | 3.52 | 39.63 | 269.07 | 5.37 | 73.83 |
| w.g. | 49 | 263.00 | 5.37 | 60.37 | 93.25 | 1.90 | 26.17 | |
| C | a.g. | 6 | 255.04 | 4.51 | 41.33 | 280.86 | 5.32 | 55.67 |
| w.g. | 49 | 313.88 | 6.41 | 58.67 | 207.63 | 4.24 | 44.33 | |
| T | a.g. | 6 | 406.79 | 7.56 | 50.73 | 65.89 | 1.22 | 50.32 |
| w.g. | 49 | 359.63 | 7.34 | 49.27 | 59.13 | 1.21 | 49.68 |
3.4 Dendrogram and cluster analysis
The UPGMA cluster analysis data showed that most of the individuals preserved by the same methods clustered together regardless of how long they were stored (Figs. 3 and 4). In the ISSR analysis of Po. tomentosa, the groups clustered into three clades with a high similarity coefficient. The first clade was formed by YI and YN, and was clustered with YD, which also clustered with YF (Fig. 3a). For S. babylonica, all individuals of each group were not able to be separated (Fig. 3b). For P. koraiensis, HF clustered with HI, which were then clustered with other groups, which were not well separated. For P. sylvestris var mongolica, the groups clustered into three clades that were made up by SF, SD and the remaining individuals. For Se. sarmentosum and H. spectabile, the groups clustered according to the different preservation methods. In RAPD analysis, for Po. tomentosa, YF and YD clustered in the first clade and YI and YN clustered in the second clade which were connected together. For S. babylonica, LF and LI formed one clade and then clustered with other individuals. For P. koraiensis, HI and HN formed one clade and clustered with HD, which then clustered with HF. For P. sylvestris var mongolica, SI and SN formed the first clade and clustered with SF, and SD clustered with the whole clade. For Se. sarmentosum, CN and CD formed the first clade and clustered with CI, which formed a sister clade with CF. For H. spectabile, the groups clustered into two clades where TF formed the first clade and other individuals formed the remaining one. In both analyses, all the fresh samples clustered together.
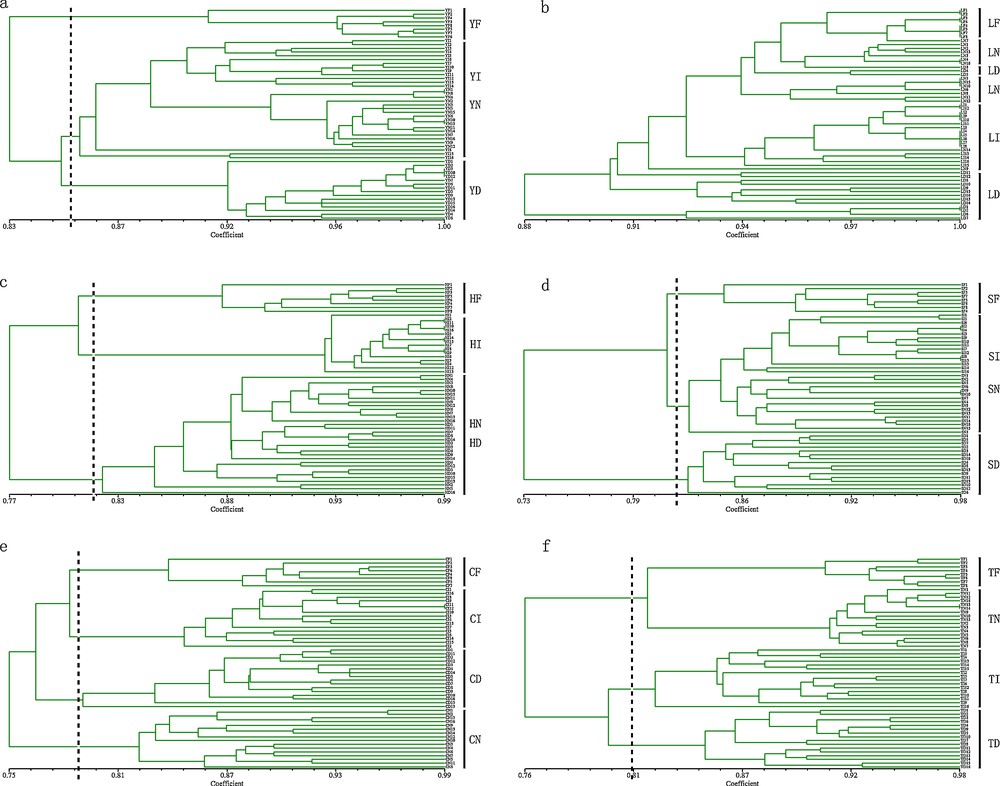
UPGMA dendrogram of six species preserved by different methods for different times based on ISSR markers. Y, Po. tomentosa; L, S. babylonica; H, P. koraiensis; S, P. sylvestris var. mongolica; C, Se. sarmentosum; T, H. spectabile; F, fresh leaves group; I, stored in −20 °C ice chest group; N, stored in liquid nitrogen group; D, dried in silica gel group; the individual's label from 1 to 8 was preserved for three weeks except for fresh groups, and from 9 to 16 was preserved for three years.
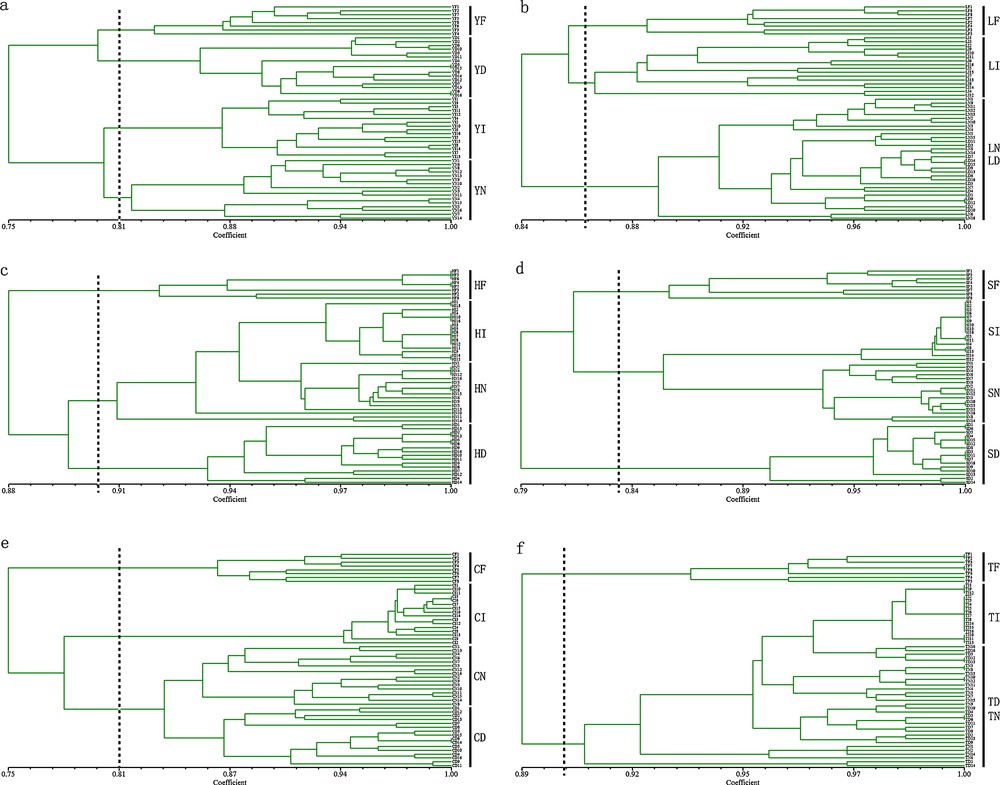
UPGMA dendrogram of six species preserved by different methods for different times based on ISSR markers. Y, Po. tomentosa; L, S. babylonica; H, P. koraiensis; S, P. sylvestris var. mongolica; C, Se. sarmentosum; T, H. spectabile; F, fresh leaves group; I, stored in −20 °C ice chest group; N, stored in liquid nitrogen group; D, dried in silica gel group; the individual's label from 1 to 8 was preserved for three weeks except for fresh groups, and from 9 to 16 was preserved for three years.
3.5 Genetic identity among different groups
In order to compare the differences between groups, we computed the genetic identity (GI) using Nei's unbiased measures based on ISSR and RAPD data (Tables 3–8). GI values between groups preserved by the same method, for different time periods ranging from 0.913 to 0.999, suggest that the same preservation methods for different storage periods had no effect on GI, both in RAPD and ISSR analyses. However, the GI values between some groups that were preserved by different methods were lower. For Po. tomentosa, the GI value between groups preserved in a −20 °C ice chest and fresh samples was 0.845 using ISSR; between groups preserved in a −20 °C ice chest and dried in silica, the result was 0.769 by RAPD. This result is lower than that observed in other groups, indicating that Po. tomentosa is not ideally suited for preservation in a −20 °C ice chest compared to other methods. Similarly, we found that drying in silica gel was not a suitable method for preserving P. koraiensis, P. sylvestris var mongolica, and H. spectabile; and storage in liquid nitrogen was not a suitable method for Se. sarmentosum. These results were consistent with cluster analysis. For S. babylonica, the GI value was more than 0.9 between all groups by ISSR analysis, and very high in RAPD analysis, indicating that the different preservation methods have little effect on the analysis.
Nei's unbiased measures of genetic identity matrix of seven groups of Po. tomentosa.
| YF | YI | YI2 | YN | YN2 | YD | YD2 | |
| YF | **** | 0.860 | 0.845 | 0.887 | 0.879 | 0.881 | 0.882 |
| YI | 0.835 | **** | 0.971 | 0.941 | 0.930 | 0.881 | 0.882 |
| YI2 | 0.833 | 0.994 | **** | 0.925 | 0.913 | 0.889 | 0.884 |
| YN | 0.886 | 0.896 | 0.896 | **** | 0.980 | 0.916 | 0.909 |
| YN2 | 0.892 | 0.871 | 0.874 | 0.988 | **** | 0.899 | 0.905 |
| YD | 0.905 | 0.769 | 0.763 | 0.872 | 0.871 | **** | 0.993 |
| YD2 | 0.908 | 0.771 | 0.766 | 0.870 | 0.876 | 0.993 | **** |
Nei's unbiased measures of genetic identity matrix of seven groups of S. babylonica.
| LF | LI | LI2 | LN | LN2 | LD | LD2 | |
| LF | **** | 0.955 | 0.936 | 0.973 | 0.972 | 0.948 | 0.941 |
| LI | 0.917 | **** | 0.981 | 0.938 | 0.941 | 0.924 | 0.915 |
| LI2 | 0.921 | 0.994 | **** | 0.934 | 0.932 | 0.930 | 0.900 |
| LN | 0.862 | 0.915 | 0.908 | **** | 0.995 | 0.951 | 0.960 |
| LN2 | 0.857 | 0.895 | 0.883 | 0.989 | **** | 0.947 | 0.953 |
| LD | 0.859 | 0.906 | 0.898 | 0.981 | 0.970 | **** | 0.961 |
| LD2 | 0.857 | 0.889 | 0.880 | 0.959 | 0.956 | 0.988 | **** |
Nei's unbiased measures of genetic identity matrix of seven groups of P. koraiensis.
| HF | HI | HI2 | HN | HN2 | HD | HD2 | |
| HF | **** | 0.869 | 0.884 | 0.882 | 0.861 | 0.838 | 0.826 |
| HI | 0.917 | **** | 0.987 | 0.867 | 0.838 | 0.819 | 0.817 |
| HI2 | 0.919 | 0.993 | **** | 0.865 | 0.842 | 0.829 | 0.830 |
| HN | 0.864 | 0.960 | 0.951 | **** | 0.975 | 0.956 | 0.942 |
| HN2 | 0.853 | 0.937 | 0.937 | 0.979 | **** | 0.977 | 0.957 |
| HD | 0.849 | 0.887 | 0.892 | 0.925 | 0.913 | **** | 0.982 |
| HD2 | 0.852 | 0.897 | 0.896 | 0.938 | 0.921 | 0.994 | **** |
Nei's unbiased measures of genetic identity matrix of seven groups of P. sylvestris var. mongolica.
| SF | SI | SI2 | SN | SN2 | SD | SD2 | |
| SF | **** | 0.912 | 0.934 | 0.901 | 0.913 | 0.814 | 0.818 |
| SI | 0.820 | **** | 0.976 | 0.946 | 0.948 | 0.850 | 0.833 |
| SI2 | 0.832 | 0.996 | **** | 0.948 | 0.959 | 0.842 | 0.837 |
| SN | 0.868 | 0.899 | 0.898 | **** | 0.983 | 0.869 | 0.864 |
| SN2 | 0.872 | 0.870 | 0.877 | 0.978 | **** | 0.876 | 0.880 |
| SD | 0.845 | 0.773 | 0.779 | 0.833 | 0.837 | **** | 0.975 |
| SD2 | 0.843 | 0.759 | 0.769 | 0.824 | 0.836 | 0.991 | **** |
Nei's unbiased measures of genetic identity matrix of seven groups of Se. sarmentosum.
| CF | CI | CI2 | CN | CN2 | CD | CD2 | |
| CF | **** | 0.890 | 0.882 | 0.860 | 0.879 | 0.878 | 0.896 |
| CI | 0.832 | **** | 0.995 | 0.863 | 0.846 | 0.861 | 0.856 |
| CI2 | 0.815 | 0.997 | **** | 0.867 | 0.850 | 0.862 | 0.852 |
| CN | 0.787 | 0.828 | 0.839 | **** | 0.965 | 0.858 | 0.845 |
| CN2 | 0.786 | 0.834 | 0.848 | 0.980 | **** | 0.861 | 0.874 |
| CD | 0.819 | 0.818 | 0.829 | 0.950 | 0.924 | **** | 0.973 |
| CD2 | 0.808 | 0.822 | 0.835 | 0.953 | 0.938 | 0.988 | **** |
Nei's unbiased measures of genetic identity matrix of seven groups of H. spectabile.
| TF | TI | TI2 | TN | TN2 | TD | TD2 | |
| TF | **** | 0.782 | 0.790 | 0.873 | 0.865 | 0.779 | 0.796 |
| TI | 0.925 | **** | 0.987 | 0.854 | 0.837 | 0.922 | 0.919 |
| TI2 | 0.918 | 0.995 | **** | 0.864 | 0.857 | 0.914 | 0.910 |
| TN | 0.936 | 0.953 | 0.948 | **** | 0.961 | 0.821 | 0.822 |
| TN2 | 0.925 | 0.967 | 0.962 | 0.998 | **** | 0.823 | 0.815 |
| TD | 0.899 | 0.967 | 0.963 | 0.941 | 0.954 | **** | 0.969 |
| TD2 | 0.900 | 0.968 | 0.964 | 0.937 | 0.950 | 0.999 | **** |
4 Discussion
Botany preservation is an important component of most studies that use ISSR or RAPD markers to estimate the extent of genetic diversity in the field. A variety of sample preservation methods have been used in the past without adequate knowledge of the effects these methods have on ISSR and RAPD analyses in various species. In this study, each storage method was used for material preservation of the three families and adequate amounts of genomic DNA for ISSR and RAPD analyses were obtained. As the materials come from the same individual of the same species, they possessed the same genetic background and should show a similar fingerprint of ISSR and RAPD markers with the low variation of bands, low genetic variation, high GI and a clustering together among groups in the same species. Moreover, if the different preservation methods had no effect on the AMOVA, an overall genetic variation of 50% should be explained by the differences among groups and within groups. If the preservation method does not fit the species, the genetic diversity may be different from the fresh groups as the DNA was not intact. However, the results showed high variation in the number of bands and PPL, and most groups preserved by different methods clustered in separate clades (Figs. 1–4, Tables 1 and 2) both by ISSR and RAPD analyses. The results demonstrate that the methods used to preserve samples have a significant effect on these analyses. In ISSR and RAPD markers, the variability or polymorphism results from the primers used, detection methods and the template DNA [23]. The high quality of the DNA should achieve credible ISSR and RAPD results; however, degraded DNA causes the number of loci to change because DNA is incomplete. For Po. tomentosa, the materials that were preserved in a −20 °C ice chest provided the least number of bands and the lowest GI compared with other groups, indicating that the method was not suitable for preserving the materials of Po. tomentosa. We have found that leaves preserved in a −20 °C ice chest quickly turn black, due to Po. tomentosa being rich in phenols, flavonoids and some readily oxidized compounds [24] that influence the extractability and integrity of genomic DNA. For S. babylonica, H. spectabile and pinaceae, the materials that were dried in silica gel obtained the least number of bands and the lowest GI compared with other groups, indicating that this method was not suitable for preserving these materials. These species have a thick epidermis, which can be 16.63 μm in P. sylvestris var mongolica [25], 11.18–17.44 μm in S. babylonica [26] and 16–20 μm in H. spectabile [27]. Most of these also possess a cuticle and overall waxiness, which gives them a good capacity to retain water and a structure that is difficult to dry in silica gel. The organic acids, enzymes, and other components in the aqueous phase can also make the genomic DNA degrade easily. Pinacea species have narrow leaves, which give them a lesser contact area with the silica gel; the leaves also provide more waxiness than Salicaceae, which make them harder to dry and preserve. We can infer that those leaves that are rich in thick, dense epidermis, cuticle or waxiness are unfit for drying in silica gel. However,as for stored in liquid nitrogen (cryopreservation), the low temperature slowed down the chemical and enzymatic reactions [28], preventing DNA degradation, making it a suitable method for preserving these materials. In contrast, storage in liquid nitrogen was not optimal for Se. sarmentosum. Se. sarmentosum possesses leaves that were recalcitrant for DNA isolation and that are rich in complex secondary metabolites such as saponin, polysaccharose, and flavone [29,30] and visible intercellular space, in addition to well-developed aerenchyma in mesophyll. Significant numbers of mucilage cells were found in the epiderm and around the vascular tissue, thus making it difficult to dry the material in silica gel and give it a strong resistance to cold stress [31]. Therefore, DNases enzyme-catalyzed reaction and chemical degradation reaction could not be immediately inactivated and resulted in DNA degradation. The different preservation methods have a disparate influence in different species on ISSR and RAPD analyses, due to the complex chemical components and specific structures of the leaves used.
We found there was no difference between samples preserved for three weeks and three years. This study has demonstrated the importance of choosing a suitable preservation method when collecting leaves in the field. We also found that the same preservation methods had differing effects on the results of ISSR and RAPD analysis. Although the two profiling techniques are based on DNA amplification by PCR, it was expected from the nature of the primers and reactions that the techniques would differ in the specific sequences targeted. The influences on ISSR and RAPD analysis by different reservation methods was mostly caused by the degradation of genomic DNA. We can infer that some other molecular markers, such as AFLP (Amplified Fragment Length Polymorphism), RFLP (Restriction Fragment Length Polymorphism), and SSR (Simple Sequence Repeats), which is based on the sufficient and high-quality genomic DNA isolated from botanical materials may also be influenced by the different preservation methods, especially for the materials come from field where far from laboratory facilities and should be stored for a few days or even long periods before they were used.
5 Conclusion
In conclusion, methods used to preserve samples from the field could have a significant effect on ISSR and RAPD analyses, and these effects vary with the species, independently of the preservation time. Our results provide a reference for researchers for selecting the most suitable preservation method depending on their study subject for the analysis of molecular markers based on genomic DNA.
Disclosure of interest
The authors declare that they have no competing interest.
Acknowledgments
We thank Dr. Wei Zhao for identifying plant species and providing the software. We also thank Dr. Ming Xing for ordering the reagents.


