1 Introduction
Genetic diversity is a fundamental component of biological diversity, and how to preserve the genetic diversity of plant species can significantly affect their long-term survival and evolution in changing environments [1]. Knowledge of the genetic diversity and population structure of protected plant species is essential for their protection and management [2–4].
Polygala tenuifolia Willd., commonly known as “Yuanzhi” in China, is a perennial herbaceous plant within the family Polygalaceae. It is an important traditional Chinese medicine (TCM) with a history of medical use for more than 2000 years. Wild P. tenuifolia is widely distributed in Shanxi, Shaanxi, Hebei, Henan, and other provinces of China, and is also distributed in Mongolia, Russia, and the Korean Peninsula at elevations of 200–2300 m [5]. Cultivation of P. tenuifolia mainly occurs in several counties of Shanxi Province. Its root bark, called Polygalae radix, is an important crude drug [6]. There is increasing commercial demand for this plant due to its excellent prevention and treatment of mental diseases (e.g., amnesia, anxiety, and insomnia) and respiratory tract ailments (e.g., asthma, bronchitis, and allergic respiratory disorders) [5]. Its medical importance generates great interest among researchers and companies to assess the existing active constituents and pharmacological effects [7,8].
However, P. tenuifolia resources are increasingly difficult to find in the wild and P. tenuifolia faces the danger of resource depletion. Slow growth has also exacerbated the depletion of wild resources [9]. Thus, it has long been listed among the Grade-III Key State-Protected Wild Medicinal Species in China [10], and strengthened protection and restoration is imminent. Most studies of P. tenuifolia have focused on its pharmacological activity [11], phytochemical properties [12], taxonomy [13], and cultivation technology [14]. Limited information is available about its genetic diversity and population structure [15], although such knowledge is fundamental to its effective conservation and sustainable utilization.
In the present study, amplified fragment length polymorphism (AFLP) analysis were employed to investigate the genetic variability in the wild and cultivated populations of P. tenuifolia. AFLP is a PCR-based marker for the rapid screening of genetic diversity and intraspecific variation without the need for prior sequence knowledge. It is a potent fingerprinting technique for genomic DNAs of any source or origin, and it can rapidly generate many highly replicable markers that allow high-resolution genotyping. Apart from AFLP, a variety of PCR-based DNA fingerprinting techniques (random amplified polymorphic DNA [RAPD], restriction fragment length polymorphism [RFLP], and simple sequence repeat [SSR]) is currently in use for molecular characterization and screening of genetic diversity [16–23], but these techniques have limitations and shortcomings, such as a lack of polymorphism (RFLP and RAPD) and the relatively low number of loci assayed (SSR) [24]. Both these limitations are removed with the use of AFLP and can be tailored to generate large numbers of polymorphisms [25]. AFLP reproducibility, reliability, resolutions, stringency, and number of loci detected per reaction do enhance its overall application. AFLP marker has largely taken over other markers for diversity, phylogenetic, and population genetics study. AFLP is very efficient in revealing polymorphisms even between closely related individuals. It has been used for recognition of individuals (paternity analysis, selfing rates, identification of cultivars, clones, etc.) and studying genetic diversity of Chinese medicinal plants [26–32].
In this study, we investigated the genetic diversity and genetic structure of P. tenuifolia. Our study aimed to:
- • reveal the level of genetic diversity in P. tenuifolia;
- • explore the distribution of genetic variation within and between the wild and cultivated populations;
- • discuss possible implications of genetic data for its management and protection.
2 Material and methods
2.1 Plant materials
We collected 382 samples representing wild and cultivated populations of P. tenuifolia from different areas of China, including Shanxi, Shaanxi, Sichuan, and Gansu Provinces. The samples were collected from 24 populations. Among these populations, six were collected from the cultivated population, 17 from the wild population and one was P. sibirica, collected as a reference substance. Collected samples were identified by Professor Wan Deguang of the School of Pharmacy, Chengdu University of TCM. The distance between the collected individual samples was at least 200 m. We collected fresh young leaves from shoots, which were dried in silica gel and stored at −80 °C before DNA was extracted. Detailed information regarding locations and codes of the study samples is shown in Table 1.
Plant materials used in the study.
| Population code | Species | Sampling localities | Sample size | Collected date |
| 1 | P. tenuifolia | Fanshi, Shanxi | 15 | July 2008 |
| 2 | P. tenuifolia | Yingxian, Shanxi | 15 | July 2008 |
| 3 | P. tenuifolia | Qinyuan, Shanxi | 15 | July 2008 |
| 4 | P. tenuifolia | Datong, Shanxi | 13 | July 2008 |
| 5 | P. tenuifolia | Fenyang, Shanxi | 15 | July 2008 |
| 6 | P. tenuifolia | Wenxi, Shanxi | 15 | July 2008 |
| 7 | P. tenuifolia | Xinzhou, Shanxi | 14 | July 2008 |
| 8 | P. tenuifolia | Wuxiang, Shanxi | 15 | July 2008 |
| 9 | P. tenuifolia | Linxian, Shanxi | 15 | July 2008 |
| 10 | P. tenuifolia | Xingxian, Shanxi | 15 | July 2008 |
| 11 | P. tenuifolia | Shouyang, Shanxi | 15 | July 2008 |
| 12 | P. tenuifolia | Xinglongshan, Gansu | 13 | July 2008 |
| 13 | P. tenuifolia | Taiyuan, Shanxi | 15 | August 2008 |
| 14 | P. tenuifolia | Wenxi (Cultivated), Shanxi | 20 | July 2008 |
| 15 | P. sibirica | Maoxian, Sichuan | 12 | July 2008 |
| 16 | P. tenuifolia | Jingxian (Cultivated), Shanxi | 20 | August 2008 |
| 17 | P. tenuifolia | Fenyang (Cultivated), Shanxi | 20 | August 2008 |
| 18 | P. tenuifolia | Maoxian, Sichuan | 15 | July 2008 |
| 19 | P. tenuifolia | Xiaxian (Cultivated), Shanxi | 20 | August 2008 |
| 20 | P. tenuifolia | Jishan (Cultivated), Shanxi | 20 | August 2008 |
| 21 | P. tenuifolia | Pucheng, Shaanxi | 15 | July 2008 |
| 22 | P. tenuifolia | Lanxian, Shanxi | 15 | July 2008 |
| 23 | P. tenuifolia | Qinyuan, Shanxi | 15 | August 2008 |
| 24 | P. tenuifolia | Xinjing (Cultivated), Shanxi | 20 | August 2008 |
2.2 DNA extraction
We extracted genomic DNA from dried leaves using a modified cetyltrimethylammonium bromide protocol [33]. The extracted DNA was detected using 0.8% agarose gel electrophoresis and stored at −20 °C until use.
2.3 AFLP fingerprinting
AFLP fingerprinting was performed in accordance with the method described by Vos et al. (1995) [24] with minor modifications. The AFLP protocol followed kit instructions (Ding Guo Biotechnology Company, Beijing China) for PCR amplification. An initial screening was performed using three individuals from wild P. tenuifolia using 64 primer combinations for selective amplifications. A total of 12 primer combinations (Table 2), which generated clear and abundant bands, were chosen for selective PCR.
Polymorphism and primer informativeness of 12 AFLP primer combinations.
| Primer combinations | Amplification bands | Polymorphism bands | PPL (%) |
| E-ACT/M-CAG | 12 | 9 | 75.0 |
| E-ACT/M-CTG | 29 | 24 | 82.8 |
| E-ACC/M-CAT | 42 | 38 | 90.5 |
| E-ACC/M-CAA | 14 | 12 | 85.7 |
| E-ACC/M-CAC | 17 | 15 | 88.2 |
| E-ACC/M-CTT | 24 | 21 | 87.5 |
| E-AGC/M-CAT | 28 | 24 | 85.7 |
| E-AGC/M-CAC | 37 | 32 | 86.5 |
| E-AGC/M-CAA | 24 | 20 | 83.3 |
| E-AGC/M-CAG | 18 | 13 | 72.2 |
| E-AGG/M-CAT | 14 | 10 | 71.4 |
| E-AGG/M-CAA | 51 | 43 | 84.3 |
| Total | 310 | 261 | – |
| Mean | 25.83 | 21.75 | 82.8 |
Amplified DNA products (4.8 μL) were mixed with 98% formamide loading buffer (1.8 μL), heated at 95 °C for 8 min and immediately cooled in an ice bath for 30 min. The resulting products were resolved by 6% polyacrylamide gel electrophoresis in 1 × Tris–borate–EDTA buffer using a 2000 bp DNA marker (TakaRa Bio, Dalian China) for 2 h at 2500 V and then 0.1% silver nitrate was used for staining.
2.4 Analysis of AFLP data
All samples were manually scored as either ‘0′ or ‘1′ corresponding to the absence or presence of AFLP bands (100–2000 bp), respectively, to construct a binary matrix. The binary matrix was edited using Microsoft Excel 2003® [34]. The percentage of polymorphic loci (PPL), effective number of alleles (Ne), Shannon's information index (I), Nei's genetic diversity (h), total gene diversity (Ht) genetic diversities within populations (Hs), coefficient of genetic differentiation (Gst), and level of gene flow (Nm) were calculated using POPGENE version 1.31 [35] with manual corrections.
The samples were treated to genetic distance and clustering analysis based on unweighted pair-group method with arithmetic means (UPGMA) using NTSYS-pc Version 2.1 [36]. Principal coordinate analysis (PCoA) was also employed to examine the genetic relationships between the detected samples using NTSYS-pc Version 2.1.
Bayesian analysis of population structures was conducted using STRUCTURE version 2.3.4 [37]. A total of 20 independent runs were performed for each set with the number of clusters (K) in the range of 2–22, a burn-in period of 1 × 104 iterations and 1 × 104 subsequent Markov Chain Monte Carlo steps. The best-fit number of groupings was evaluated using ΔK [38] in Excel 2003®.
3 Results
3.1 Genetic diversity
Among 64 AFLP kit primer pairs, 12 pairs amplified well-distributed fragments with good distinctness, which were highly polymorphic and with size range of 100–2000 bp. A total of 310 clear and quantifiable fragments was generated in 24 populations. Among 310 loci, 261 (84.19%) were polymorphic. The total number of fragments of each primer combination was in the range of 12–51, with an average of 25.83. The percentage of polymorphism was 71.4–90.5%, with an average of 82.8% per primer combination. The primer E-ACC/M-CAT had the highest gene polymorphism, with PPL = 90.5%; and primer E-AGG/M-CAT had the lowest polymorphism, with PPL = 71.4% (Table 2).
In the combined data matrix of all 12 primers, h and I were 0.3296 and 0.4822, respectively. The genetic diversities within populations (Hs) and within species (Ht) were 0.2831 and 0.3235, respectively (Table 3). The coefficient of genetic differentiation (Gst) between populations was 0.1250. Based on the Gst value, Nm was estimated as 3.4989 (i.e. Nm > 1). These results indicated high gene flow and low differentiation between the wild and cultivated populations.
Genetic diversity and differentiation parameters of P. tenuifolia.
| Source level | PPL (%) | N e | h | I | H t | H s | G st | N m |
| Wild | 81.9 | 1.5538 | 0.3154 | 0.4635 | – | – | – | – |
| Cultivated | 63.9 | 1.4375 | 0.2507 | 0.3688 | – | – | – | – |
| Populations | 84.2 | 1.5834 | 0.3296 | 0.4822 | 0.3235 | 0.2831 | 0.1250 | 3.4989 |
| St. Dev | – | 0.3538 | 0.1775 | 0.2440 | 0.0324 | 0.0273 | – | – |
At the population level, the wild population (PPL = 81.9%, h = 0.3154, I = 0.4635) showed a higher genetic diversity level than the cultivated population (PPL = 63.9%, h = 0.2507, I = 0.3688; Table 3).
3.2 Genetic distance and structure
The genetic distance and structure of P. tenuifolia were analyzed based on AFLP markers using NTSYS-pc and STRUCTURE. The STRUCTURE analysis based on the ΔK method revealed that ΔK = 31.19 for K = 3, and ΔK < 3.86 for all other values of K (2 and ranging within 4–21) (Fig. 1). The optimal ΔK for K = 3 showed that the best-fit model for the sampled 23 populations of P. tenuifolia revealed three clusters: the first two groups were the wild one, and the other was the cultivated one (Fig. 2). The UPGMA-based phenogram (Fig. 3) showed that the 24 populations were divided into two groups at 0.67: the first one was P. tenuifolia and the other was P. sibirica. This result was consistent with the morphological classification [5]. The genetic distance coefficients ranged from 0.37 to 0.67. The 23 P. tenuifolia populations were divided into two groups: group I was the wild population and II was the cultivated population (Fig. 3). This corresponded to the results of STRUCTURE analysis based on a Bayesian approach. The existence of three groups was supported by the PCoA (Fig. 4). The 24 populations were divided into two groups firstly: the first one was the cultivated from population 14, 24, 16, 17, 20 and 19, and the other one was comprised of the wild populations. The wild P. tenuifolia populations were clustered into two groups further: the first one consisted of population 1 to 4, and the other one was made up of other populations. Two-dimensional PCoA separated the 24 populations into three distinct clusters, distinguishing the wild and cultivated populations. The three-dimensional PCoA differentiate P. sibirica from P. tenuifolia (Fig. 5).
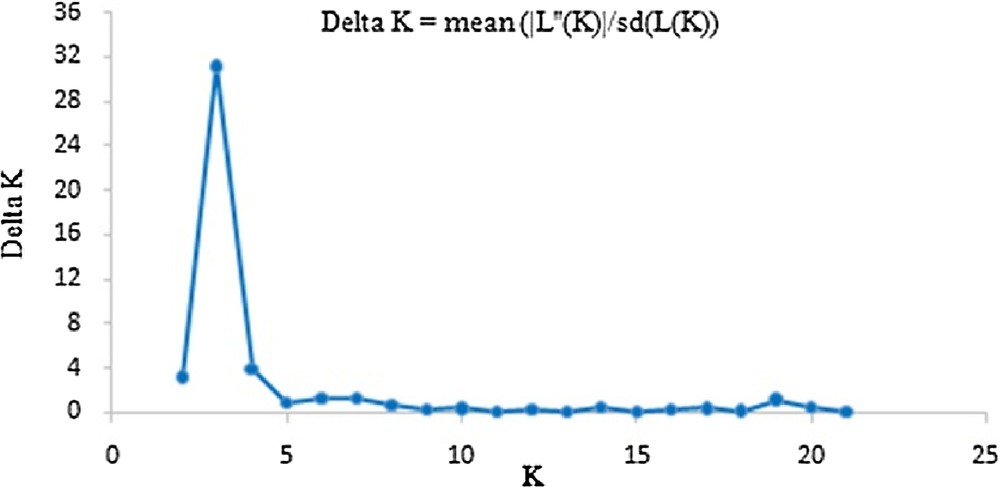
ΔK values from the mean log-likelihood probabilities from STRUCTURE runs where the inferred clusters (K) ranged from 2 to 21.
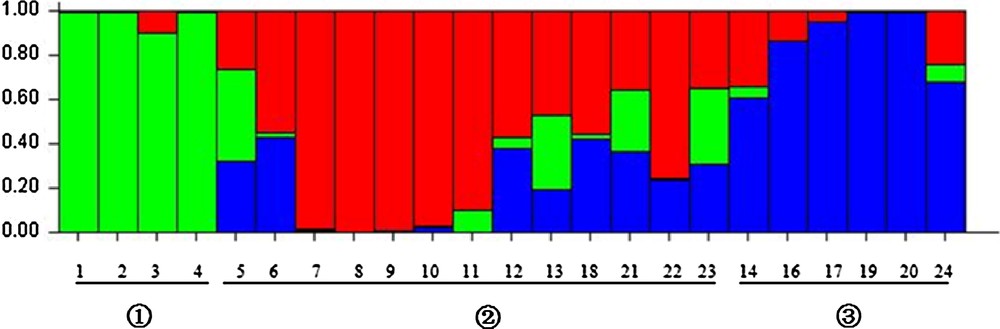
Estimated genetic clustering (K = 3) obtained with the STRUCTURE program for 23 populations (not included No. 15). Individuals are separated according to the population, and the black vertical line in the bar chart is the population identifier.
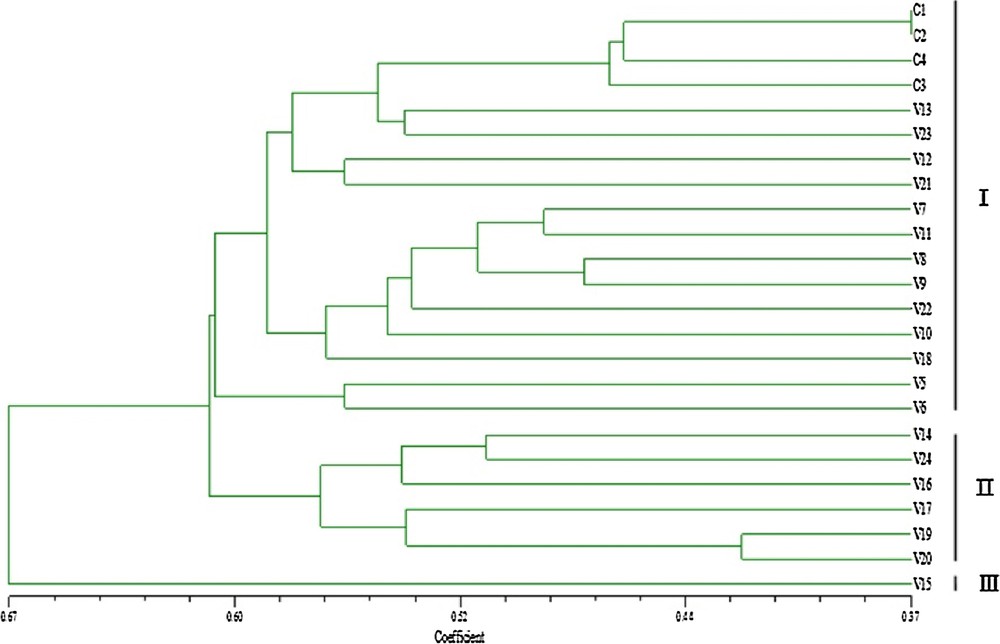
Phenogram of 24 populations based on unweighted pair-group method with arithmetic means (UPGMA).
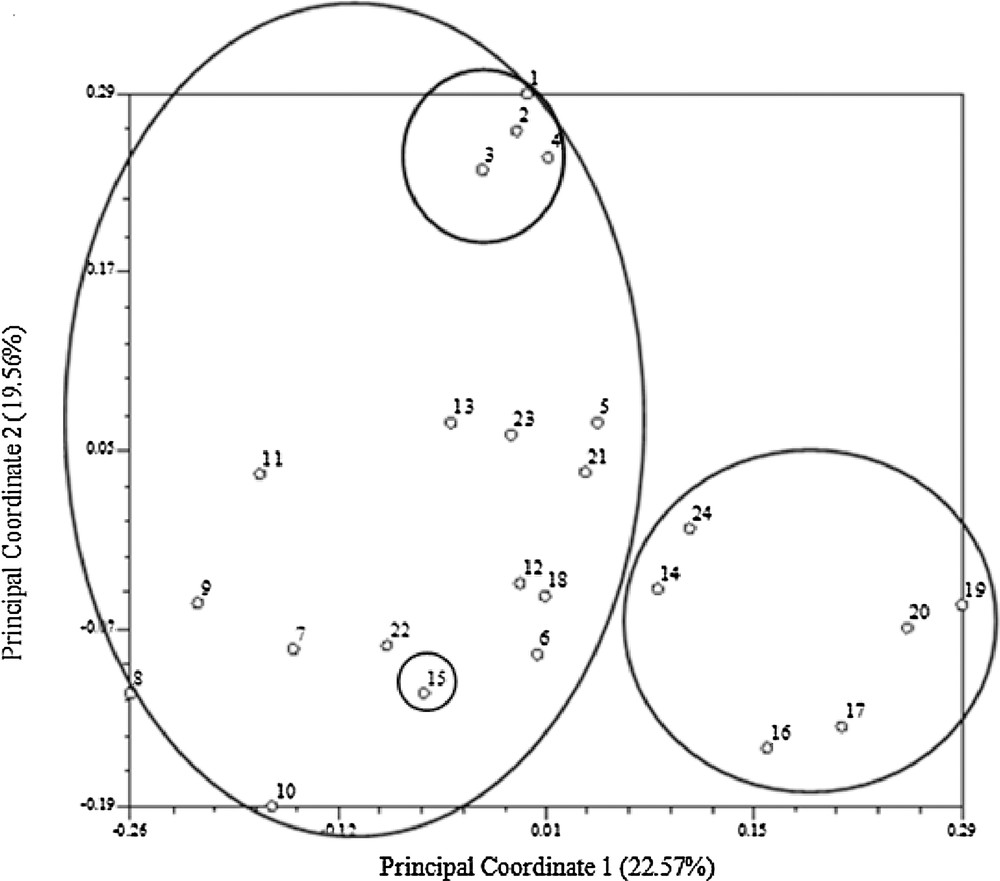
Two-dimensional graph produced from the principal coordinate analysis of 310 AFLP loci of 24 populations.
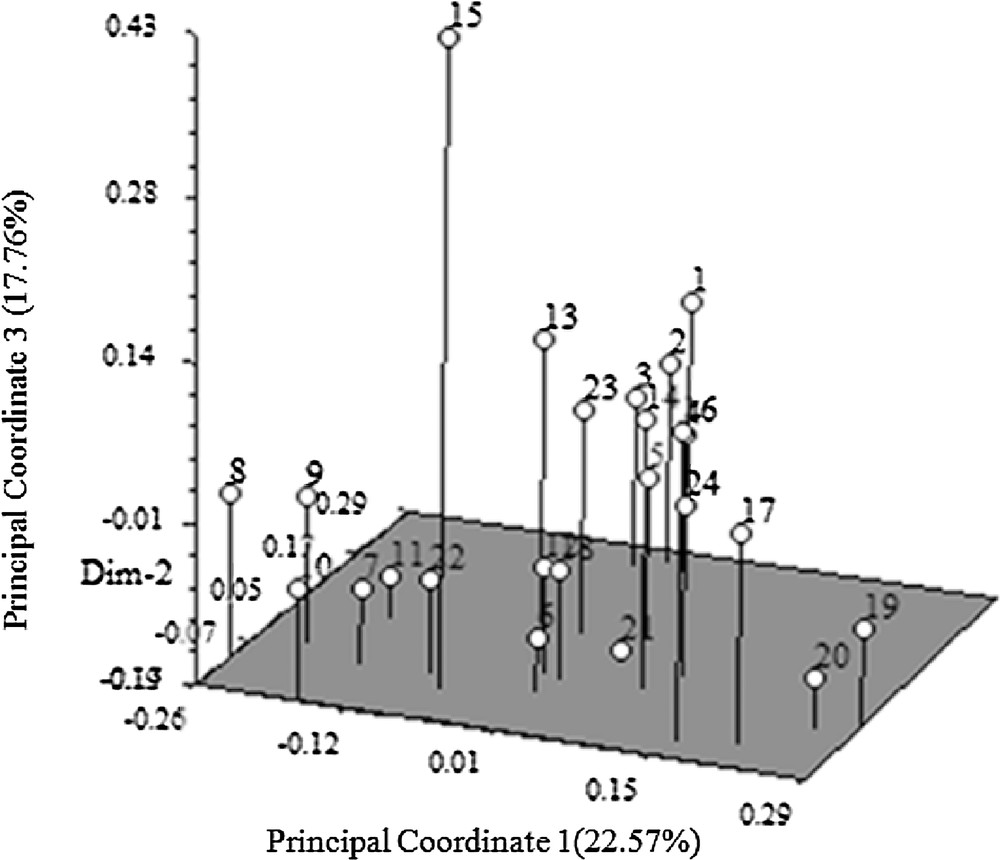
Three-dimensional graph from the principal coordinate analysis of 310 AFLP loci of 24 populations. (Dim-2: principal coordinate 2 [19.56%]).
4 Discussion
4.1 Genetic diversity
The analysis of AFLP marker data in 23 populations of P. tenuifolia, representing three sampled populations, revealed high genetic diversity in the species (PPL = 84.2%, h = 0.3296, I = 0.4822). The results of the present study were comparable with those of another study of P. tenuifolia [15]. They made similar estimates of genetic diversity (PPL = 90.9%, h = 0.3100, I = 0.4620) at the species level, but GST = 0.7448 was higher and Nm = 0.1713 was lower than those of the present study (with GST = 0.1250 and Nm = 3.4989) because of different selected populations.
In the present study, P. tenuifolia had a high genetic diversity at the species level. Genetic diversity and population structure of a species is correlated with its geographic distribution, life form, breeding system, seed dispersal, gene flow, natural selection, and habitat fragmentation [39]. A species with wide distribution, long lifespan, frequent gene flow and large quantities of seeds usually has high genetic diversity [40]. Although the reproductive biology of P. tenuifolia has not been studied in detail, its capsules are small, with wing-like structures that allow them to be dispersed by wind and the seeds are collected by ants, both of which will contribute to gene diversity. In addition, P. tenuifolia is a perennial herb with wide geographical distribution, consistent with high genetic diversity. In contrast to the high level of genetic variability in the wild P. tenuifolia, the genetic diversity in the cultivated population was relatively moderate. The genetic diversity of species in large populations is higher than in small populations because of genetic drift and inbreeding [41,42]. The cultivated P. tenuifolia survives in the farm, its genetic diversity often decreases because there may be a close relative of pollination, or due to geographical constraints.
High genetic variation enables species to adapt to changing environments [43]. The presence of moderate to high genetic diversity in the two populations indicated that the current resource depletion of P. tenuifolia is not caused by genetic factors such as genetic diversity decline, genetic drift, and inbreeding. This result is similar to that for the endangered species Rhododendron protistum var. giganteum [44]. The main threat to P. tenuifolia may be ecological and human factors, which include habitat destruction and over-exploitation. Although we did not conduct a detailed habitat survey, the limited individuals found in the wild may partly support this hypothesis. However, the factors that lead to the resource depletion of the wild P. tenuifolia should be further elucidated.
4.2 Genetic structure
The clustering of the AFLP data clearly separated the 382 individuals into wild and cultivated populations, excepting P. sibirica, using NTSYS-pc. This was consistent with analysis of genetic structure using STRUCTURE software and was also supported by the PCoA. Gene flow is a critical determinant of population genetic structure. The Gst and Nm between the two extant populations were 0.1171 and 3.7686, respectively; and the high Nm was congruent with the low Gst among P. tenuifolia populations. Research has revealed that geographic isolation and domestication play important roles in the evolution of the genetic structure. There was geographic isolation between the wild and cultivated populations in the present study, but the gene flow was high, possibly because P. tenuifolia has only been cultivated on a large scale for 30 years in the south of Shanxi Province. Most perennial crops like P. tenuifolia show fewer features of the domestication syndrome than annual seed crops, and domestication may occur more slowly because fewer sexual generations occur in a given period of time [45]. Farmer-led domestication in P. tenuifolia has resulted in a slight reduction in genetic variation in the cultivated compared with wild populations at the AFLP markers. Artificial selection during domestication often results in the narrowing of the genetic base of the cultivated populations. However, perennial domesticated species usually retain relatively higher percentages of the variation of their wild ancestors compared with annual plants [45–47]. For example, AFLP markers showed that populations of the cultivated tropical fruit tree Spondias purpurea retained approximately 90% of the variation detected in wild populations [48]. The reduction in genetic diversity observed for cultivated P. tenuifolia was similar to the reductions seen in other long-lived species. Thus, farmer-led domestication activities likely reduced the genetic diversity of cultivated P. tenuifolia in Shanxi; however, this did not greatly affect diversity within populations.
4.3 Genetic diversity and phenotypic diversity
The clustering of the AFLP data clearly separated these individuals into P. tenuifolia and P. sibirica groups using NTSYS-pc. P. tenuifolia leaves are linear to linear-lanceolate, the racemes are terminal, the flower keel is glabrous abaxially, and the stamens filaments form an open staminal ciliate sheath split in upper 1/4 with two separate single filaments, and its capsules are eciliate. By contrast, P. sibirica lower leaves are ovate, the upper leaves are lanceolate or elliptic-lanceolate, the racemes are extra-axillary or pseudoterminal, the flower keel is hairy abaxially, the stamen filaments form an open ciliate staminal sheath united at least in lower 2/3, and its capsule are shortly ciliate. The phenotypic variation was consistent with this study genetic classification [5].
4.4 Implications for conservation and sustainable use
Our study of the genetic diversity of P. tenuifolia has important implications for the conservation and management of the resource depletion of the species. Undoubtedly, in situ conservation is the most effective method to conserve species resources [49]. For P. tenuifolia, a widely distributed species, representative units of conservation should be defined in each climatic zone for in situ conservation. The high genetic diversity in P. tenuifolia indicates that the major factors threatening the persistence of plant resources are ecological and human factors (e.g., habitat destruction and over-exploitation) rather than genetic. Considering the sustainable use of these resources, we suggest that management policies be improved to maintain an appropriate effective population size of P. tenuifolia and to protect its natural habitats. As a perennial wild herb, the wild resources will be rapidly consumed by years of unrestrained mining. Setting the excavation time interval to limit the amount consumed aims to increase the number of wild individuals. Since P. tenuifolia is also cultivated, a good conservation strategy for wild resources should involve the substitution of cultivated for wild plants. There is a similar set of implications for the conservation of Rheum tanguticum [50]. Both wild and cultivated populations of P. tenuifolia are valuable gene pools for conservation and future breeding. Farmers must be trained to maintain good collection practices of currently cultivated P. tenuifolia germplasm as they regenerate the species in subsequent cycles of mating and selection in on-farm propagation [51]. Since the heritability of important characters, including root bark traits, is unknown, we also recommend establishing progeny trials with seed collected from all representative units that should include cultivated populations to protect the resources of different genotypes. We believe that the threatened state of P. tenuifolia will be soon relieved when the above strategies are implemented.
5 Conclusion
In conclusion, the genetic diversity and structure of P. tenuifolia were examined in this study using AFLP markers, and we detected high genetic diversity at the species level, and high gene flow and low genetic differentiation between the wild and cultivated populations. These results showed that the major factors threatening the persistence of P. tenuifolia resources are ecological and human factors rather than genetic. Our hope is that these results will help design species conservation and management programs, such as in natural habitat conservation, setting the excavation time interval for resource regeneration and the substitution of cultivated for wild plants.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grant numbers 31270381, 2012).


