1 Introduction
Arsenic (As) is one the most toxic metals and its contamination occurs due to natural processes as well as human activities [1]. This toxic metalloid can find its way to water bodies causing drinking water contamination. Arsenic, therefore, can find its way in many foods such as rice, which is a staple food in many countries [2]. Long-term As exposure leads to arsenicosis that causes major health problems such as cancers of different body parts [3]. Arsenite [As(III)] and arsenate [As(V)] are the most predominant forms of As, and As(III) is known to be 100 times more toxic than As(V) [4]. Arsenic is a metal that is toxic for all life forms, including plant, animal, bacteria, and human [5].
Many microorganisms have the ability to resist and detoxify As and other such toxic metals through mechanisms such as exclusion [6], compartmentalization [7], formation of complexes [6], biosorption [8], and enzymatic detoxification [9]. The genes responsible for As resistance and detoxification are located in the ars operon [10]. On the other hand, biosorption is a physiochemical process of removing toxic heavy metals from environment with the help of living organisms [11]. Biosorption utilizes dead or living biomass to chelate heavy metals from aqueous solutions. In this process, sorbate is any metal ion or molecule that is to sorb on a sorbent that is any solid surface, thus forming a sorbate–sorbent complex. A variety of functional groups is present at the bacterial cell surfaces that serve as binding sites for metallic ions [12].
Purple non-sulphur bacteria (PNSB) are one of the most diverse group of bacteria, capable of adapting a variety of metabolic life styles depending on the type of nutrients available in their habitat and on the presence or absence of oxygen as well as light. They can grow photoheterotrophically in anoxic light environment, oxidizing various organic carbon sources while respiring difference chemical compounds, thus reducing them in the process. Many PNSB are also known to resist and detoxify many toxic metals and elements such as As. PNS bacteria are also known to produce hydrogen when cultured anaerobically in the presence of incandescent light, which is a clean and renewable fuel and is widely accepted as a potential substitute for fossil fuels [13].
The objective of the present study was to check PNSB tolerance towards As and to determine whether these bacteria can detoxify As by respiring it. PNSB could be one of the ideal organisms for the bioremediation of As and such toxicants due to their versatile metabolic nature [14].
2 Materials and methods
2.1 Samples collection
Two samples were collected separately. One sample was taken from Quaid-e-Azam Industrial Estate, Kot Lakhpat, Lahore, Pakistan. The other sample was taken from paddy fields, University of the Punjab, Lahore, Pakistan. The samples were collected in such a way that they contained both water and the corresponding sediments. Both samples were taken from a depth of 2–2.5 feet.
2.2 Enrichment and isolation
Arsenic-resistant PNSB were enriched by inoculating the samples in a Mineral Salt Succinate (MSS) broth [15] containing 1.0 mM of As(V) (Na2HAsO4·7 H2O). The medium was prepared anaerobically in 500-mL Schott bottles and the enrichment cultures were incubated under continuous incandescent light of ∼ 3000 lux at 30 °C (Memmert model ICP, Germany) for two weeks. After incubation, different serial dilutions of the enrichment cultures were plated on the MSS medium both with and without As(V). The plates were incubated in an anaerobic jar (OXOID AG0025, England) containing an anaerobic sachet (Thermo Scientific Oxoid AnaeroGen, England) placed under continuous incandescent light at 30 °C for two weeks. Morphologically diverse colonies were picked and purified by several rounds of quadrant streaking. Gram reaction, catalase, and oxidase tests were performed. All experiments were performed in triplicates if not mentioned otherwise.
2.3 Determination of metal resistance in the isolates
An MSS broth containing 1.0, 2.5 and 5.0 mM of As(III) and 1.0, 2.5, 5.0 and 10 mM As(V) was prepared separately. In the case of Cu, Hg, Co, Ni, Se, Pb, and Cr, a concentration of 0.5 mM was used in the MSS broth. The tubes were inoculated with fresh MSS broth cultures anaerobically and aseptically. The tubes were placed in continuous incandescent light at 30 °C for 1 week.
2.4 Determination of As-respiration
MSS agar was prepared and the carbon source, i.e. sodium succinate, was replaced with other carbon sources such as citrate, oxalate, lactate, acetate, and propionate. Such media were prepared both with and without 0.5 mM As(V). The medium was inoculated with the bacterial isolates and incubated in anaerobic jars under continuous incandescent light (3000 lux) at 30 °C for 1.0 week.
2.5 Estimation of pigment production
2.5.1 Estimation of photopigment
The MSS broth was inoculated with a fresh bacterial culture and incubated under anaerobic conditions. The bacterial cells were harvested at 1844 g for 20 min. The supernatants were discarded, and the pellets were resuspended in bovine serum albumin with 0.25% w/v. Absorbance was taken at 495 nm, 591 nm, and 864 nm with an UV–Vis spectrophotometer (CECIL CE 7200, Aquarius, UK).
2.5.2 Estimation of dry cell weight & carotenoid content
Bacterial cells were harvested by centrifugation at 6869 g for 20 min at 4 °C (Sigma 2K30 Laborzentrifugen, Germany) and resuspended in a phosphate buffered saline 1.0X at pH 7.4. Each cell suspension was divided into two sets by shifting half of the cell suspensions into fresh tubes. The cells were harvested by centrifuging them again. Supernatants were discarded, and pellets were washed thrice with autoclaved water. One set of pellets was dried by placing them in a hot-air oven at 105 °C for 24 hours. The dry cell weight was calculated by the following formula taken from Al-Azad, Soon, and Ransangan [16]:
dry cell weight (g·L−1) = (final weight − initial weight)/V × 1000
where
initial weight = weight of the tube (g)
final weight = weight of the tube (g) + dry pellet weight (g)
V = volume of the sample (mL).
The other set of pellets was resuspended in an acetone–methanol solution (2:7 v/v). The suspensions were covered with an aluminium foil and incubated at 4 °C for 30 min. Following this the suspensions were centrifuged again and the optical density of the supernatants was measured at 480 nm using an UV–Vis spectrophotometer (CECIL CE 7200, Aquarius, UK). The carotenoid content was calculated using the formula given by Jalal et al. [17]:
C = D × V × F (2500)/dry weight of the sample (g)
where
C = total carotenoid content (mg·g−1)
D = absorbance at 480 nm
V = total volume of the sample used
F = dilution factor of the sample
2500 = average extinction coefficient for carotenoids.
2.6 Determination of As oxidation/reduction
A bacterial inoculum was prepared aerobically in an MSS broth. The cells were harvested by centrifugation at 1428 g. The pellets were resuspended in 10 mL of 0.09% (v/v) normal saline. Then the cell suspension was distributed in different tubes that were then supplemented with 0.5 mM of As(V) or 0.1 mM of As(III). Controls were also set up that were devoid of any As. Half of the tubes were incubated aerobically while the other half was incubated anaerobically, both for 5 h at 30 °C. After incubation, the suspensions were further divided into two equal halves. The suspensions were centrifuged at 1844 g for 10 min. Supernatants and pellets of one half of the tubes were separated and stored at −20 °C till further analysis. The pellets of the other half of the tubes were resuspended in 0.5 M EDTA. EDTA cell suspensions were again centrifuged at 1844 g for 10 min, and the supernatant was then transferred to fresh tubes and stored at −20 °C till further analysis.
2.6.1 Qualitative analysis of As oxidation/reduction
As oxidation/reduction in the above-mentioned stored samples was determined following the method of Salmassi et al. [18]. One hundred and sixty-six microliters of each sample was added in the wells of a microtiter plate along with 5 μL of 0.01 M KMnO4, and colour change was observed. Negative controls (not containing any arsenate) and positive controls (arsenate standards of known concentrations) were also used. Purple to yellowish colour change indicates As(V) reduction. The selected isolates were investigated further for extent of As oxidation/reduction through quantitative As assay.
2.6.2 Quantitative analysis of As oxidation/reduction
The extent of As oxidation/reduction was estimated following the method of Cummings et al. [19] with minor modification, as reported by Shafique et al. [20]. Absorbance was taken at 865 nm using a spectrophotometer (CECIL CE 7200, Aquarius, UK). The standard curve was made by treating the standards similarly. The concentration of oxidized/reduced As was calculated using the formula of the standard curve.
2.7 Detection of hydrogen gas production
For detection of hydrogen gas production, the syringe displacement method was used [21]. For this purpose, an MSS broth medium was prepared with some modifications. The medium contained 0.33 g of KH2PO4, 0.33 g of MgSO4·7 H2O, 0.33 g of NaCl, 0.5 g of NH4Cl, 1.0 g of sodium succinate, 20 mg of yeast extract, 0.5 g of K2HPO4, 0.4 g of l-glutamine, 1.0 mL of trace salt solution, 500 μL of 0.0001% (v/v) FeSO4·7 H2O, and 1.0 mL of vitamin solution per litre. The solution of vitamins contained 10 mg of biotin, 35 mg of niacin, 30 mg of thiamine dichloride, 20 mg of p-aminobenzoic acid, 10 mg of calcium pentothenate, 5 mg of vitamin B12, 10 mg of pyridoxine hydrochloride per 100 mL of distilled water. The solution of vitamins was sterilized by filtration and added to the medium aseptically after autoclaving. The medium was prepared anaerobically and dispensed in autoclaved serum bottles aseptically. The bottles were sealed with rubber stoppers and aluminium crimps. The inoculum was prepared in LB-broth. Syringe needles (3 mL) were inserted in serum bottles through the rubber stoppers aseptically so that the tip of the needle was dipped in the medium. The bottles were incubated under continuous incandescent light at 30 °C for 1.0 week. The displacement of the syringe plungers was taken as an indication of gas production.
2.8 SSU rRNA gene sequencing and phylogenetic analysis
PCR was performed to amplify the SSU rRNA gene by using 27f (AGAGTTTGATCCTGGCTCAG) and 1522r (AAGGAGGTGTGCCARCCGCA) primers. The selected isolates were sequenced for SSU rRNA gene from Macrogen Inc. (South Korea). The sequences obtained by this Sanger sequencing method were checked for base call quality through Finch TV and were cleaned. The NCBI BLAST tool was used to find homology between the isolates and other bacteria.
2.9 Statistical analysis
Statistical analysis was performed in Microsoft Excel 2013. The results were displayed as the mean values and the standard deviation was displayed as error bars.
3 Results
3.1 Isolation, characterization and identification of As-resistant PNSB
The temperature of the sample taken from Quaid-e-Azam Industrial Estate was 34 °C, and its pH was 8.0. The temperature of the sample taken from paddy fields, University of the Punjab, was 29 °C; its pH was 7.5. Five isolates were selected based on arsenic resistance, reduction potential, and biosorption. Three of the isolates (Q3C, Q3B, and Q2B) belonged to Quaid-e-Azam Industrial Estate, Kot Lakhpat, Lahore, Pakistan, whereas the rest of the two isolates (PFI and PFII) were from paddy field, University of the Punjab, Lahore, Pakistan. Maroon colonies of PNSBs were obtained on the MSS agar plates, which were then purified by many rounds of quadrant streaking. All the isolates were gram-negative rods, and catalase and oxidase positive. SSU rRNA gene sequence analysis and NCBI BLAST results revealed the identity of Q3B as Rhodospirillum species (95% homology) and of Q3C as Rhodospirillaceae sp. (98% homology). The sequences were submitted in Genbank under accession numbers MH807574 and MH807575 for Q3B and Q3C, respectively.
3.2 Resistance of toxic metals in PNSB
Rhodospirillum sp. Q3B, Rhodospirillaceae sp. Q3C and PFII showed resistance up to 10 mM As(V). Rhodospirillaceae sp. Q3C was able to resist 5 mM As(III) (Table 1). Se and Ni (0.5 mM) were resisted by all five isolates. Co (0.5 mM) was resisted by all isolates except isolate H1. All isolates showed resistance to Pb (0.5 mM), except Rhodospirillaceae sp. Q3C. Cu (0.5 mM) was resisted by all the isolates, except Rhodospirillaceae sp. Q3C and isolate H2 (Table 2).
Arsenic resistance by PNSB.
| Bacterial isolates | Arsenic resistance | ||||||
| As(V) 1.0 mM |
As(V) 5.0 mM |
As(V) 10 mM |
As(III) 0.5 mM |
As(III) 1.0 mM |
As(III) 2.5 mM |
As(III) 5 mM |
|
| Rhodospirillum sp. Q3B | +++ | +++ | +++ | +++ | +++ | − | − |
| Rhodospirillaceae sp. Q3C | +++ | +++ | ++ | +++ | +++ | ++ | ++ |
| PFII | +++ | + | + | +++ | +++ | + | + |
| H1 | +++ | − | − | − | +++ | − | − |
| H2 | +++ | + | − | + | − | − | − |
Resistance to toxic metals by PNSB.
| Bacterial isolates | Hg 0.5 mM |
Pb 0.5 mM |
Ni 0.5 mM |
Se 0.5 mM |
Cr 0.5 mM |
Co 0.5 mM |
Cu 0.5 mM |
| Rhodospirillum sp. Q3B | − | ++ | + | ++ | − | ++ | ++ |
| Rhodospirillaceae sp. Q3C | − | − | + | ++ | − | +++ | − |
| PFII | − | ++ | + | + | − | ++ | + |
| H1 | − | + | + | ++ | − | − | +++ |
| H2 | − | + | + | + | − | + | − |
3.3 As-respiration by PNSB
Anaerobic fermentation of the carbon sources by the bacterial isolates was analysed in the absence of As. Acetate was fermented by Rhodospirillaceae sp. Q3C, PFII and isolate H2, as growth was observed by these bacteria under anaerobic conditions in media containing only this carbon source. Lactate was fermented only by PFII. Citrate and oxalate were fermented by Rhodospirillum sp. Q3B and PFII, while propionate was fermented by Rhodospirillum sp. Q3B, PFII and H2 (Table 3).
Carbon source utilization and As-respiration by PNSB.
| Strains | Carbon sources Fermentation [without As(V)] | Carbon sources Respiration [with As(V)] | ||||||||
| Acetate | Lactate | Citrate | Propionate | Oxalate | Acetate | Lactate | Citrate | Propionate | Oxalate | |
| Rhodospirillum sp. Q3B | − | − | − | + | ++ | ++++ | +++ | ++++ | +++ | + |
| Rhodospirillaceae sp. Q3C | ++ | − | − | − | − | ++++ | ++ | ++++ | + | +++ |
| PFII | ++ | + | ++ | ++ | ++ | ++++ | ++ | ++ | ++++ | ++ |
| H1 | − | − | − | − | + | − | +++ | ++++ | ++ | + |
| H2 | + | − | − | − | − | ++++ | + | + | − | + |
Those isolates that did not ferment different carbon sources were provided with As(V) to check whether these bacteria can use these carbon sources as electron donors and As(V) as electron acceptor, thus respiring As. All the isolates were able to respire As(V) in the presence of lactate, citrate, and oxalate, except PFII. All the isolates except H1 were able to respire As(V) in the presence of acetate. In the presence of propionate, all the isolates showed As(V) respiration except isolate H2 (Table 3).
3.4 Photopigment production by PNSB
All isolates showed maximum absorption at 864 nm, indicating the presence of bacteriochlorophyll a [22] (Table S1).
3.5 Dry cell weight and carotenoid content measurements
Rhodospirillum sp. Q3B, Rhodospirillaceae sp. Q3C and PFII showed 8.33, 1.67 and 16.6 g·L−1 of dry cell weight, respectively. Rhodospirillaceae sp. Q3C was found to contain highest carotenoid content up to 5.6 mg·g−1. Isolate PFII contained 0.18 mg·g−1 and Rhodospirillum sp. Q3B contained 1.1 mg·g−1 of carotenoid content (Table S1).
3.6 As oxidation/reduction by PNSB
In qualitative assays, all the isolates showed colour change from purple to yellowish brown, indicating the reduction of As(V) to As(III). The colour change was more pronounced in anaerobic conditions as compared to aerobic conditions. In quantitative assays, Rhodospirillaceae sp. Q3C was found to show 7.92% (v/v) biosorption aerobically and 11.675% (v/v) anaerobically. As(V) reduction by this bacterium was 47.42% (v/v) aerobically and 76.675% (v/v) anaerobically. Rhodospirillum sp. Q3B showed 14.43% (v/v) biosorption aerobically and 19.27% (v/v) anaerobically. This bacterium showed 56.67% (v/v) As(V) reduction aerobically and 74.92% (v/v) As(V) reduction anaerobically (Fig. 1).
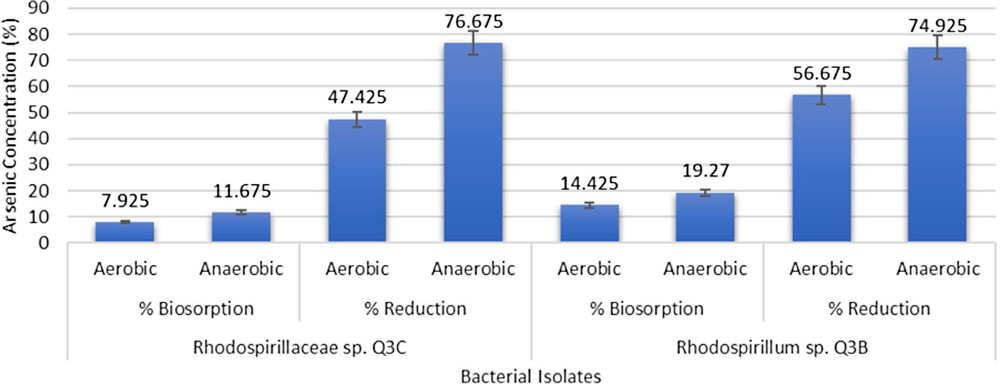
Arsenic reduction and biosorption by Rhodospirillum sp. Q3B and Rhodospirillaceae sp. Q3C.
3.7 Detection of hydrogen gas production by PNSB
Rhodospirillum sp. Q3B and Rhodospirillaceae sp. Q3C showed gas production under anaerobic conditions in the presence of incandescent light, as indicated by the displacement of syringe plungers. No gas production or displacement of the syringe plunger was detected in the control serum bottles (Fig. 2).
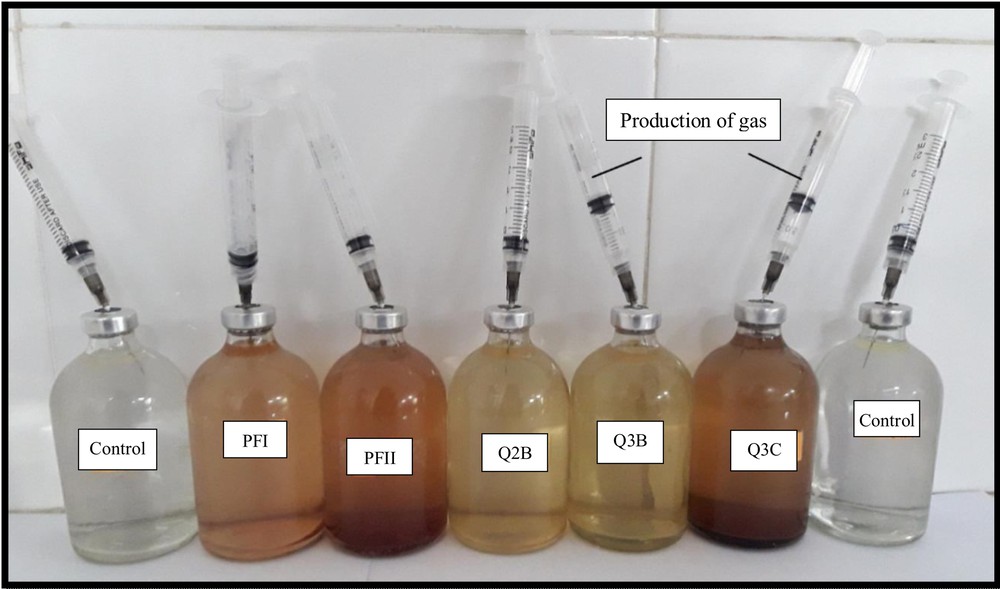
Hydrogen gas production demonstrated by purple non-sulphur bacteria (PNSB) isolates Rhodospirillum sp. Q3B and Rhodospirillaceae sp. Q3C under anaerobic conditions.
4 Discussion
Being metabolically diverse, PNSB hold great potential as tools for biotechnology purposes such as wastewater treatment, bioremediation, and bio-hydrogen production. They can prove a very inexpensive way to deal with many environmental problems as they can easily grow in a variety of environments such as rice fields, industrial effluents, marine water, coastal lagoons, fish ponds, etc. [23,24]. Metal-resistant PNSB have potential use for remediation of heavy metal contaminated sites [2,25]. The present study focuses on the ability of PNSB to detoxify and respire As along with photopigment, carotenoid, and hydrogen production.
Arsenic detoxification by the bacteria was initially determined by qualitative method using KMnO4 in which colour changes from purple to yellowish brown indicated reduction of As(V) to As(III). The colour change was more pronounced in anaerobic samples of Rhodospirillaceae sp. Q3C, Rhodospirillum sp. Q3B and H1. These isolates were further analysed for the extent of As- reduction and biosorption [9]. Bacteria-mediated metal reduction is a process in which bacteria transforms the valence state of the metal, affecting its toxicity. This is followed by extrusion of the metal out of the bacterial cell [6]. Rhodospirillaceae sp. Q3C was highly efficient and reduced 76.67% (v/v) As(V) in anaerobic conditions, while in aerobic conditions Rhodospirillum sp. Q3B reduced the highest As(V) concentration up to 56.67% (v/v). Bacteria possess this reduction potential due to the presence of ars operon and other related detoxification mechanisms [26]. Batool et al. [27] reported 62.9% (v/v) As(V) reduction by R. palustris strain CS2 (6.29 ± 0.24 mM). These purple non-sulphur bacteria were also found to resist other heavy metals. Sakpirom et al. [28] reported R. palustris strain TN110 to reduce Cd up to 84.21 ± 0.28% (v/v) and Zn up to 54.83 ± 0.70% (v/v).
Many bacteria are known to oxidize organic compounds anaerobically by reducing As, a process that is called As-respiration [29]. Many As-transforming PNSB of the present study were found to grow anaerobically only when provided with As(V). Thus these bacteria obtained energy by respiring As(V). Huber et al. [30] reported that Pyrobaculum arsenaticum (type strain PZ6), a strict anaerobic archaeon isolated from hot spring, utilizes As(V) as a terminal electron acceptor in respiration. Yamamura et al. [31] reported dissimilatory arsenic reduction by Bacillus sp. strain SF-1. The bacterium was also found to reduce selenate under anaerobic conditions. Though many bacteria are known to respire arsenic, reports of PNSB involved in arsenic respiration are, however, scarce. Nookongbut et al. [32] reported possible arsenic respiration by Rhodopseudomonas palustris strain C1.
Maximum carotenoid content was observed in Rhodospirillaceae sp. Q3C up to 5.6 mg·g−1. There is an indirect correlation between dry cell weight and carotenoid content. Heat from light source is trapped by high cell density, thus affecting the carotenoid content as it is heat sensitive [33]. Jalal et al. [34] reported highest carotenoid content by Rhodopseudomonas sp. IIUM-JHR2 up to 4.08 ± 2.21 mg·g−1 after three days of incubation.
The bacteria were identified by SSU rRNA gene sequence classification via NCBI BLAST. Q3B showed 95% homology with Rhodospirillum rubrum. R. rubrum is an important PNSB that has showed significant potential in hydrogen and bioplastic production, as mentioned by Reslewic et al. [35]. Q3C showed 98% homology with Rhodospirillaceae species. The family Rhodospirillaceae is composed of 18 species placed in the four genera Rhodopseudomonas, Rhodospirillum, Rhodocyclus, and Rhodomicrobium, many of which are known to produce hydrogen [36].
Under anoxygenic light conditions, the gas produced by PNSB is hydrogen gas [37]. PNSB have the ability to produce hydrogen gas when suitable conditions and environment are provided. Bianchi et al. [21] reported Rhodopseudomonas palustris AV33 as one of the best candidates to produce hydrogen gas (up to 1.2 L per litre of culture). Basak and Das [38] reported Rhodobacter sphaeroides strain O.U.001 to produce hydrogen in bioreactor up to 4.5 ± 0.05 mol. Rhodopseudomonas palustris, with some mutation in its gene that constitutively express nitrogenase genes, has the ability to produce specifically hydrogen gas when supplied with media containing ammonium as a nitrogen source [39,40]. In PNSB, hydrogen production occurs by process called photofermentation that utilizes the breakdown of small-chain fatty acids into H2 and CO2 [41]. The enzyme responsible for hydrogen production in PNSB is nitrogenase. In the absence of nitrogen, the nitrogenase enzyme of PNSB is fully dedicated to H2 production [42]. However, minor amounts of other gases such as CO2 can also be produced when PNSB perform photofermentation [43].
In the light of all above-mentioned experiments, it is clear that metabolically diverse PNSB hold potential solutions for environmental problems. The present study utilizes PNSB for As reduction and adsorption along with oxidation of organic compounds as well as hydrogen production. This gives a comprehensive view of how such diverse bacteria can be used to achieve multiple goals simultaneously. Hydrogen gas produced by these bacteria can be used for energy purposes. However, the production of hydrogen needs to be further evaluated and estimated at different conditions. Moreover, these processes need to be operated and optimized simultaneously to achieve maximum results along with field trials.
Contribution of the authors
HS and QUK contributed equally in the experimental work. HS drafted the manuscript. YR planned and supervised the work as well as he reviewed and finalized the manuscript.
Compliance with ethical standards
The current study does not involve any experiments performed on human beings or animals.
Funding
Funding to carry out this research was provided by the University of the Punjab, Lahore, Pakistan.
Research involving human participants and/or animals
Not applicable.
Informed consent
Not applicable.
Disclosure of interest
The authors declare that they have no competing interest.
Acknowledgements
The University of the Punjab, Lahore, Pakistan, is acknowledged for providing funds for performing this research work.


