1. STING, a central protein of the immune response associated with cytosolic nucleic acids
The innate immune system is the first line of defence against pathogens. One of the central pathways for pathogen detection is the recognition of their nucleic acid content by intracellular receptors. A large panel of receptors involved in the initiation of the innate immune response has been described [1]. Their interaction with pathological nucleic acids leads to the activation of signalling pathways that induce the production of inflammatory cytokines. This inflammatory response mobilises effector cells of the adaptive immune system and is therefore a crucial step in the initiation, maintenance and coordination of immune responses.
Recently, a central pathway for the detection of cytosolic nucleic acids has been described (Figure 1). This pathway was originally identified in immune cells as activated by cytosolic double-stranded DNAs during viral or bacterial infections [2]. This pathway relies on the cyclic GMP-AMP synthase (cGAS) receptor and the Stimulator of Interferon Genes (STING) adaptor protein. The cGAS receptor recognises cytosolic nucleic acids, including double-stranded DNA (dsDNA), single-stranded DNA (ssDNA) or DNA:RNA hybrids [3, 4, 5]. The interaction of cGAS with cytosolic nucleic acids induces its activation, enabling its capacity to synthesize the 2′3′-cGAMP cyclic dinucleotide from ATP and GTP [6]. This secondary messenger interacts with the adaptor protein STING, promoting the assembly of a signalosome in which STING serves as an anchoring site for the various partners required for the induction of the inflammatory response. Those partners notably include the protein TANK binding kinase 1 (TBK1) and the interferon regulatory factor 3 (IRF3) and/or nuclear factor κB (NF-κB) transcript factors [7, 8]. Activation of the cGAS-STING signalling axis is characterised by the production of type I interferons, besides other pro-inflammatory cytokines and chemokines. Thus, this signalling pathway is particularly critical in the response to pathogens, as the production of type I interferons leads to the establishment of an antiviral state through induction of the expression of interferon response genes.
Overview of the cytosolic DNA sensing via the cGAS-STING pathway. dsDNA presence in the cytosol can result from pathogen infection or self-DNA after cellular stress. cGAS binds dsDNA leading to cGAS activation and synthesis of 2,3′-cGAMP from ATP and GTP. cGAMP binds to ER-resident STING, this binding will trigger STING translocation to the Golgi apparatus. In the Golgi, STING binds to and activates TBK1 (by autophosphorylation) as well as the transcription factor NF-κB via IKK. TBK1 then phosphorylates IRF3 which undergoes dimerization and translocates to the nucleus to induce transcription of type I IFNs and several inflammatory chemokines. NF-κB induces transcription of inflammatory cytokines. Created with Biorender.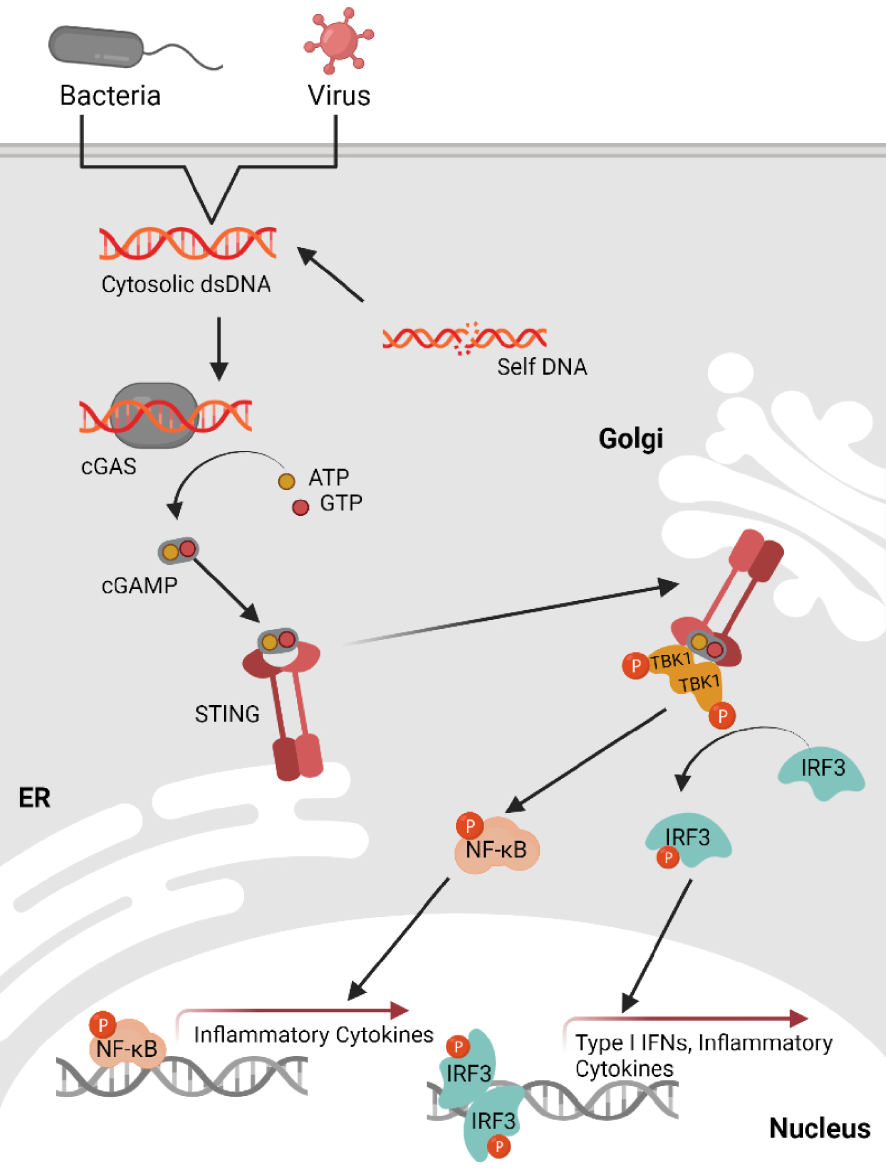
Recent work suggests that cGAS and STING beyond their canonical function in innate immunity, are also involved in cellular processes such as the induction and maintenance of senescence or the regulation of cell proliferation [9, 10, 11].
2. STING in fatty acid metabolism: an unexpected role
In 2022, we revealed a central role of STING in energy and glucose homeostasis via the regulation of adipose tissue metabolism [12] (Figure 2). Using a mouse model deleted for the STING protein [13], this study showed that STING inhibits the fatty acid desaturase 2 (FADS2) enzyme essential for the desaturation of dietary polyunsaturated fatty acids (PUFAs). Therefore, ablation or agonist-induced degradation of STING increased FADS2 activity, leading to the accumulation of PUFA derivatives and driving thermogenic gene activation. As a result, white adipose tissue showed signs of a browning process. Notably, white adipose depots were metabolically more active, burning energy through adaptive thermogenesis. PUFAs were also shown to bind STING and inhibit its activity. Modulation of PUFA production thus regulates STING-dependent antiviral responses while contributing to the resolution of STING-associated inflammation. Finally, this study also revealed that STING agonists can bind and modulate the enzymatic activity of FADS2. Thus, this work demonstrates the existence of a negative regulatory loop between STING and FADS2, opening many questions pertaining to their respective roles in human pathologies presenting with a chronic STING-associated inflammation [14].
STING plays a role in lipid metabolism. Convergent data in 2022 showed that STING regulates lipid metabolism. In Drosophila, STING form a multi-protein complex with two enzymes involved in long-chain fatty acid synthesis FASN and ACC. In mouse STING regulates the activity of the lipid desaturase FADS2 modulating the synthesis of long-chain polyunsaturated fatty acid. The production of those lipid species will in turn modulate STING activity and narrow its activation in presence of its agonist cGAMP. In human populations several alleles exist and were already known to be more or less potent in the induction of IFN response. The study of AQ and HAQ alleles showed that they also differ in lipid metabolism modulation as HAQ promotes more fat storage than AQ. Created with Biorender.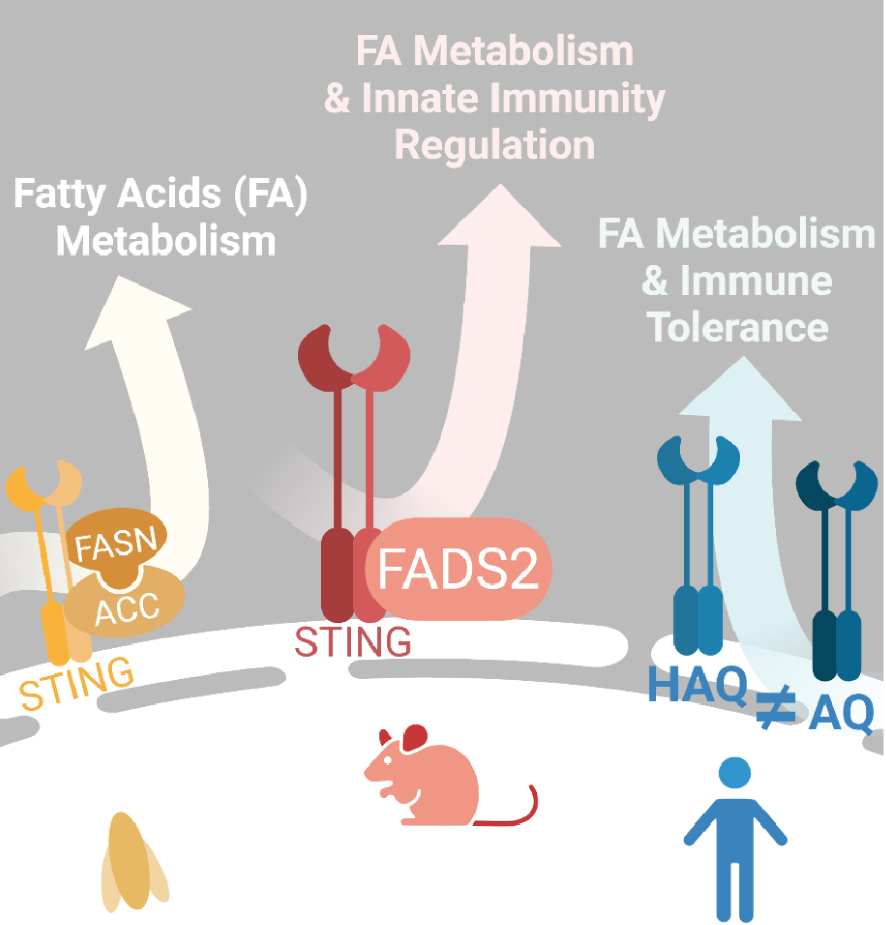
Interestingly, alleles of STING not competent for the induction of interferon responses have been documented in human populations. These include the STING HAQ allele that bears three amino acid substitutions: R71H-I229A-R292Q [15]. Analysis of STING haplotypes revealed a high prevalence of the HAQ allele in East Asian populations as compared to sub-Saharan African populations. Conversely, sub-Saharan African populations show a high prevalence of an AQ allele (G230A, R293Q) competent for the recognition of cyclic dinucleotides and the induction of interferon responses [16]. The authors propose that these STING alleles (HAQ and AQ) underwent natural selection during migration out of Africa 50,000–70,000 years ago [16]. Using mouse models expressing the HAQ or AQ human STING alleles, they showed that AQ mice store less lipid than HAQ mice, while displaying improved fatty acid oxidation. Furthermore, the visceral white adipose tissue of HAQ mice was infiltrated by regulatory T cells and anti-inflammatory macrophages, unlike that of AQ mice. Thus, the HAQ haplotype promotes fat storage, while the AQ haplotype promotes immune tolerance. As the HAQ allele confers better energy storage capacity than the AQ allele, they conclude that this ability to store energy could have been critical during ice age for the survival of early humans [17]. The study by Vila et al. showed that the HAQ allele of STING strongly interacts with FASD2 [12], which is consistent with the description of a prominent role of this allele in fatty acid metabolism [16]. Thus, the study of the evolution of STING haplotypes suggests that the function of STING in modulating fatty acid metabolism may have been critical during human migration out of Africa.
In support of this hypothesis, work in Drosophila [18] confirmed the conservation of the functional link between STING and fatty acid metabolism. The authors showed that the Drosophila STING orthologue interacts with enzymes involved in the biosynthesis of long-chain fatty acids such as acetyl-CoA carboxylase (ACC) and fatty acid synthetase (FASN) within a multiprotein complex. Furthermore, the absence of STING in Drosophila led to decreased lipid storage and decreased expression of genes related to lipid metabolism. Thus, the absence of STING increased sensitivity to food deprivation and oxidative stress in Drosophila. These results showing profound lipid metabolism alteration in Drosophila support a conserved role for STING in the regulation of lipid metabolism.
Finally the FADS (FADS1, FADS2 and FADS3) genomic region was found to be under selection in Greenlandic Inuit, probably driven by a diet high in PUFAs [19]. Those results confirming FADS genetic selection among populations [20] suggest that these genes play an important role in human adaptation to diets. Thus the study of STING and FADS allele co-evolution could bring new insight into their synergistic role in inflammation and metabolic functions.
3. Regulation of lipid metabolism by STING: a link to innate immune regulation?
Although the role of STING in host defence mechanisms against pathogens is well established, these recent findings on the link between STING and fatty acid metabolism demonstrate that this protein has important functions beyond the detection of cytosolic nucleic acids. Evolutionary analysis of STING suggests that the modulation of lipid metabolism is ancient, conserved and may be the primordial function of STING.
Metabolic pathways that interact with STING, such as those leading to the production of PUFAs, are also involved in the control of STING-independent inflammatory responses. Indeed, while PUFAs can directly inhibit STING [12], they can also be processed into lipid mediators (oxylipins) that possess, among others, immunomodulatory properties [21]. Thus, it can be hypothesised that STING alleles expressed in organisms lacking an interferon signalling pathway could participate to the control of inflammatory responses by regulating the generation of these lipid metabolites.
Altogether, these recent studies [12, 16, 18] highlight a tight link between innate immunity and lipid metabolism, paving the way for research into the role of metabolic alterations in human pathologies associated with aberrant STING activation [22]. Those notably include cancer, where a crucial role of a functional cGAS-STING axis has been recently shown to be essential for driving tumour immunogenicity [23, 24]. The role of STING-dependent metabolic regulation remains to be investigated in this context.
Conflicts of interest
Authors have no conflict of interest to declare.




 CC-BY 4.0
CC-BY 4.0