1. Introduction
The origin of the eukaryotic cell, eukaryogenesis, represents a major evolutionary transition (“in individuality”), together with the origins of life and of complex multicellular organisms [1, 2]. Eukaryogenesis resulted in a significant increase of average cell complexity as compared to generally simpler, prokaryotic, cells. This process was followed by a rapid and wide diversification of lineages. Most extant eukaryotic diversity corresponds to unicellular organisms, generally called protists, but several lineages include multicellular organisms with more or less specialised tissues, such as animals, plants, fungi or kelp, among others [3, 4, 5] (Figure 1). This phylogenetic radiation was accompanied by the adoption of different life strategies. Although most eukaryotes are phagotrophic organisms that can engulf smaller prey, some of them acquired photosynthesis and yet others, such as fungi or oomycetes (Stramenopiles), became osmotrophic and, often, parasitic, secreting enzymes for the extracellular digestion of complex molecules [3, 4]. Through these different lifestyles, and having in general bigger cells, eukaryotes colonised brand new ecological niches, and contributed to increasing the complexity of trophic networks in ecosystems [6, 7].
Scheme of the tree of eukaryotes showing the phylogenetic relationships among major eukaryotic groups and supergroups. Eukaryotic supergroups (indicated by different colors) usually encompass many different phyla and represent major ancestral splits in the eukaryotic tree. Drawings illustrate examples of the type of organisms included in the corresponding taxa. The vast majority of eukaryotic diversity correspond to unicellular, mostly flagellated and heterotrophic, protists. Eukaryotic photosynthesis evolved by the endosymbiosis of the cyanobacterial ancestor of plastids at the onset of Archeplastida, which are primarily photosynthetic, and subsequently spread to other lineages by secondary endosymbiosis. Clades including photosynthetic members with primary plastids (directly derived from their common cyanobacterial ancestor) and secondary plastids (derived from endosymbiotic red or green algae) are indicated by colored circles. Groups comprising complex multicellular lineages are also indicated: Opisthokonta comprise animals, fungi and their unicellular relatives; Chloroplastida, green algae and plants; Rhodophyta, red algae; and Stramenopiles, brown algae and many unicellular photosynthetic and heterotrophic lineages. The rest of the groups comprise mostly unicellular lineages (or possessing simple filamentous or colonial multicellularity). The classical supergroup “Excavates” may not be monophyletic; it comprises the Discoba (including the photosynthetic euglenids and diverse heterotrophic protists) and Metamonada (including well-known parasites, such as Giardia and Trichomonas spp. and several free-living anaerobic protists). The “CRuMs” is a relatively recently established clade that includes several heterotrophic flagellated protists often displaying filopodia. The Amorphea include, in addition to the well-known Opisthokonta, the Amoebozoa (classical amoeba), the heterotrophic flagellated apusomonads and the anaerobic/microaerophilic amoeboflagellated breviates. In addition to green algae and plants (Chloroplastida) and red algae (Rhodophyta), Archaeplastida include glaucophyte algae, which encompass a handful of low-abundance species found in freshwater ecosystems. Cryptista group the photosynthetic cryptophyte algae as well as heterotrophic flagellates. Haptista is a recently recognized clade that encompass centrohelid heliozoans and haptophyte algae, some of which, such as Emiliania huxleyii, have calcifying exoskeletons and are abundant in oceans. The TSAR supergroup includes telonemid flagellates and the classically recognized SAR clade: Rhizaria (radiolarians, foraminiferans, cercozoans—including some photosynthetic species—, and related lineages), Stramenopiles (widely diverse lineages of heterotrophic nanoflagellates but also of unicellular—diatoms, golden algae, bolidophytes—and multicellular—phaeophytes or brown algae—algae) and Alveolata (ciliates, photosynthetic and heterotrophic dinoflagellates, and related lineages). The position of the root of the eukaryotic tree is not reliably known (indicated by a multifurcation at the base of the tree).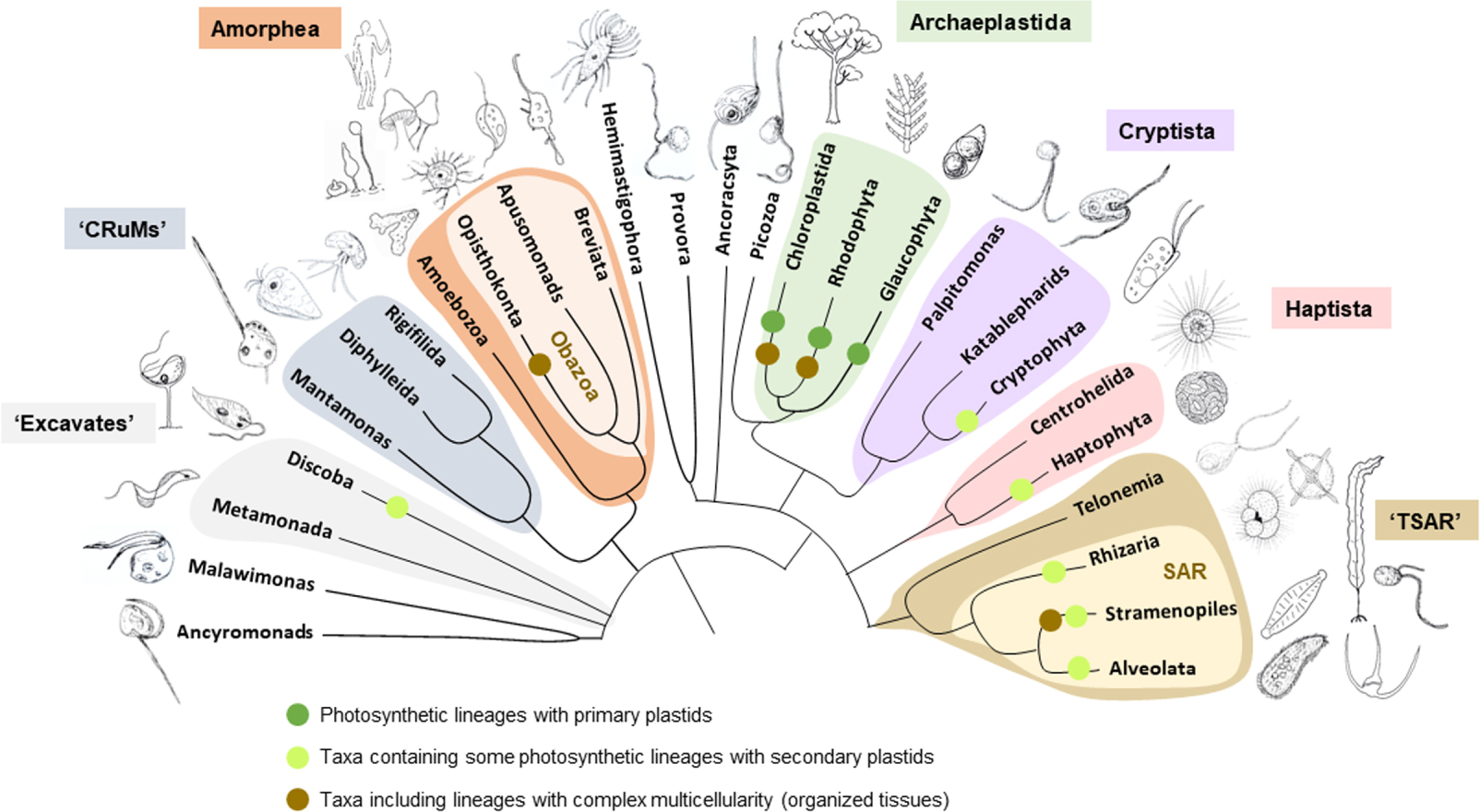
In contrast to complex multicellularity, which emerged independently several times among eukaryotes [3, 5], the origin of the eukaryotic cell was unique, i.e. it occurred only once during the evolution of life on Earth. Although the root of the eukaryotic tree remains to be robustly placed [5] (Figure 1), molecular phylogenetic analyses show that all extant eukaryotes form a monophyletic group in the tree of life [8], and share a same last eukaryotic common ancestor (LECA) [9, 10]. Comparative genomics allows to infer an already quite complex LECA, which looked pretty much like a modern eukaryote. With far more than 5000 genes [9], LECA already had high metabolic flexibility and complexity, possessing a developed cytoskeleton and an endomembrane system with a nucleus and nuclear pores, a complex trafficking network (Golgi apparatus, endosomes, etc.), an endoplasmic reticulum, and endo- and exocytic pathways. LECA was also endowed with flagella, ubiquitin signalling and proteasomes, an intron splicing system, RNA interference, mitosis and cytokinesis, and most likely meiosis and sex [9, 11]. LECA also possessed mitochondria [12]. Although some eukaryotic lineages including Microsporidia and diplomonads (then referred to as Archezoa) were initially thought to be truly amitochondriate and appeared to branch deeply in the eukaryotic tree [13], they were subsequently shown to have mitochondrial derivatives and branch within other eukaryotic clades with more robust methods of phylogenetic inference [12, 14]. Consequently, LECA was in all points comparable to modern eukaryotes.
How did the complex eukaryotic cell originate? In this short, far from exhaustive, review, we will briefly examine how early ideas on eukaryogenesis, which were largely based on autogenetic processes, transformed over time to increasingly recognise the crucial role of microbial symbiosis and have now become openly symbiogenetic. We will discuss the impact that the discovery of a particular group of prokaryotes, the Asgard archaea, had in this conceptual shift. Finally, we will comment on current challenges to discriminate between existing eukaryogenetic models and build a consensual detailed model of eukaryogenesis.
2. Early ideas on the origin of eukaryotes
2.1. From autogenous models to a three primary domain view
The discovery and study of microbial life has always been linked to the development of technological tools, the first of which was, obviously, the microscope. Since Antonie van Leeuwenhoek’s times, progress in optical microscopy had led to distinguish different microbial forms, which incidentally helped Ernst Haeckel to reconstruct the first universal phylogenetic tree based on morphological traits [15]. Photonic microscopy allowed to distinguish two very broad types of cells, one cell type with rather simple morphology and usually small size (generically called bacteria) and another type with typically larger size and widely diverse, sometimes decorated, forms (including notably protozoa and microalgae). In the first decades of the twentieth century, electron microscopy allowed to show that those two cell kinds were underlain by distinct ultrastructure. In 1937, Chatton coined the terms prokaryotic and eukaryotic for them [16], albeit the final delimitation of prokaryotes and eukaryotes dates only from the 1960s [17, 18]. The defining feature of eukaryotes was the nucleus (etymologically from the Greek eu, true, good, and , core, kernel). In eukaryotes, DNA replication and transcription occur in the nucleus while translation occurs in the cytoplasm; by contrast, in many prokaryotic cells transcription and translation are often coupled, i.e. protein synthesis can start as mRNA is being synthesized.
Under this dichotomy of cell structural types, most early models envisaged that eukaryotic cells evolved from prokaryotic cells (bacteria) by a process of complexification. This process would be autogenous, i.e. involving the development of already existing elements or the evolution of new ones in an originally prokaryotic cell. One example of such models is that proposed by Tom Cavalier-Smith in 1975, whereby not only the endomembrane system and the nucleus, but also membranous organelles such as mitochondria and plastids, would derive from thylakoids in a cyanobacterial-like ancestor of eukaryotes [19]. Much less popular, the idea that prokaryotes evolved by reduction of a eukaryotic-like ancestor of extant life was also proposed [20, 21, 22]. However, this option left the putative origin of eukaryotic-level complexity from pre-cellular systems fully unexplained. The initial autogenous bacteria-to-eukaryote transition models appeared linear (Figure 2A), but were soon to be challenged by the discovery of a so-far overlooked group of organisms, the archaea, and its impact on the tree of life.
Evolution of concepts about eukaryogenesis. (A) Early models whereby eukaryotes would result from the complexification of simpler bacteria. (B) Models based on the existence of three primary domains whereby eukaryotes would derive from a proto-eukaryotic lineage sister to archaea that developed complex features, including the nucleus, endomembranes and phagocytosis, prior to the late acquisition of the alphaproteobacterial ancestor of mitochondria. (C) Current favored models whereby eukaryotes derive from the symbiotic merging of one Asgard-related archaeon and one or more (dotted line) bacterial partners. In this view, only archaea and bacteria are primary domains; eukaryotes form a third, but secondary, domain of life.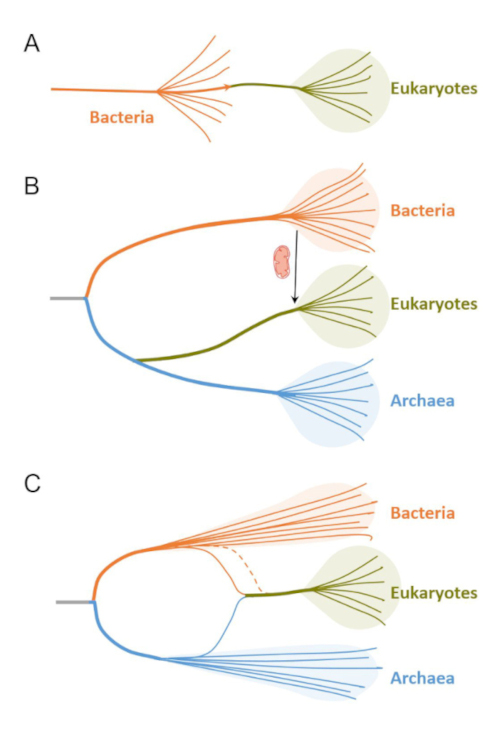
The discovery of archaea was a consequence of the so-called “molecular revolution” that took place during the second half of the twentieth century and has not ceased to develop until today. Any biological classification system must be based on phylogenetic relatedness. Based on morphological standards, bacterial species were difficult or impossible to distinguish from one another, and for a long time microbiologists used numerical taxonomy relying on various phenotypic traits of cultured clonal species. However, these traits do not allow the establishment of evolutionary relationships among bacterial taxa, such that Roger Stanier and collaborators pessimistically concluded in 1963 that “the ultimate scientific goal of biological classification cannot be achieved in the case of bacteria” [23]. Fortunately, at about that time, it became clear that evolutionary information could be stored in the nucleotide or amino acid sequence of biological polymers [24], paving the way for the development of molecular phylogeny. Based on this and with the aim of establishing a natural phylogenetic classification of all extant life, including bacteria, Woese and colleagues were the first to use conserved molecular markers (small subunit rRNA) to reconstruct a universal phylogenetic tree [25, 26]. Surprisingly, a clade of bacteria-looking organisms, which mostly thrived in extreme environments (hot and/or acidic springs, solar salterns) or were methanogenic, segregated from both classical bacteria and eukaryotes. This clade was first called Archaebacteria [25] before the official definition of this group as a third domain of life, the Archaea, together with those of Bacteria and Eucarya [26]. The discovery of archaea posed a new problem for the origin of the eukaryotic cell, as now there were two structural cell types but three different phylogenetic domains of life. Where did eukaryotes come from?
Interestingly, in the first rooted universal phylogenetic trees, archaea appeared as sisters to eukaryotes [26, 27, 28]. Some authors even found a higher similarity of some archaea to eukaryotes based on ribosome comparisons [29]. Furthermore, this sister relationship was consistent with the progressive realisation that archaeal “informational” genes and processes, i.e. involved in DNA replication and repair, transcription and translation, resembled more (or only) their eukaryotic counterparts [30, 31]. Accordingly, the scenario that emerged and has been mainstream until recent years, postulated that eukaryotes evolved from a third proto-eukaryotic lineage that shared a more recent common ancestor with archaea (Figure 2B). This third primary lineage would have developed all typical eukaryotic features except for the mitochondrion, including an endomembrane system, the nucleus, a developed cytoskeleton and, notably, phagocytosis [32, 33]. Phagocytosis would have been crucial for the acquisition of the mitochondrial ancestor, which was shown around the same time to derive from ancient endosymbiotic bacteria.
2.2. The symbiotic origin of mitochondria and plastids
The idea that some membranous organelles in eukaryotes evolved from endosymbiotic bacteria dates back to the early twentieth century. In 1905, Konstantin Mereschkowsky suggested that plastids derived from “blue-green algae” (Cyanobacteria) [34, 35] and in 1927, Wallin proposed that mitochondria derived from purple bacteria (Alphaproteobacteria) [36]. However, those ideas remained controversial and faded away from stage until 1967, when Lynn Margulis (then known as Lynn Sagan) popularised them again [37], making a case for the importance of symbiosis in eukaryotic evolution [38]. Mitochondria and plastids did not form de novo, but divided from existing organelles and bore features that might as well be remnants of a previous free-living bacterial stage, such as their own genome and bacterial-like ribosomes [37]. The definitive demonstration of a bacterial origin for these organelles came from molecular phylogenetic analyses of organellar genes (coxI, rbcL), which unambiguously placed mitochondria as sister to Alphaproteobacteria and chloroplasts as sister to Cyanobacteria [39].
The cyanobacterial endosymbiosis at the origin of plastids had major evolutionary and ecological consequences, as it led to the evolution of photosynthetic eukaryotes. The primary endosymbiosis with the plastid ancestor was shown to be unique and gave rise to the Archaeplastida, which encompasses three major clades: glaucophyte algae, red algae, and green algae and land plants [40, 41]. Subsequently, secondary (and even tertiary) endosymbioses of red and green algae within a not yet fully established number of heterotrophic eukaryotic hosts gave rise to additional lineages of photosynthetic eukaryotes [3, 42, 43] (Figure 1). For a long time, the specific cyanobacterial lineage at the origin of the plastids remained elusive. However, it has recently been shown that the plastid ancestor was related to the newly discovered clade of the Gloeomargaritales [44, 45]. This cyanobacterial order encompasses unicellular species thriving exclusively in freshwater or terrestrial ecosystems, with a marked preference for hot springs, which suggests that eukaryotic photosynthesis evolved in continental systems [44, 46]. A second independent event of primary endosymbiosis leading to a stable symbiosis with a high level of integration, albeit not as achieved as that of plastids, is also known. It involved a member of the Synechococcus–Prochlorococcus clade within a testate amoeba of the genus Paulinella [47].
Like in the case of plastids, with time and more mitochondrial sequences from diverse eukaryotes, it soon appeared clear that mitochondria were monophyletic and descended from a unique mitochondrial ancestor related to Alphaproteobacteria, already present in LECA and likely a facultative aerobe [12]. The transition from free-living bacterium to organelle involved, like in plastids, the transfer of many genes to the eukaryotic nuclear genome and the implementation of targeting pathways for the import of host-produced products into the mitochondrion [48]. This reductive process was more drastic in anaerobic protists, where mitochondria lost aerobic respiration, and in some parasitic lineages, which completely lost the mitochondrial genome and retained only minimal functions, notably iron–sulphur cluster assembly [49, 50]. The alphaproteobacterial lineage at the origin of mitochondria remains elusive. Although affinities with the Rickettsiales, which notably encompass intracellular parasitic bacteria, and the marine Pelagibacterales were proposed, they resulted from phylogenetic reconstruction artefacts induced by a similar convergent compositional bias, namely high AT-rich genomes. More robust analyses, including a better taxonomic sampling and more accurate methods of phylogenetic reconstruction, currently place mitochondria out of the known alphaproteobacterial lineages [51, 52].
2.3. Early symbiogenetic models of the eukaryotic cell
For the last part of the twentieth century and until recently, the prevailing scenario of eukaryogenesis invoked a third proto-eukaryotic lineage sister to archaea that, at some point, acquired mitochondria by endosymbiosis. However, several models started to question the existence of such a primary lineage for which direct evidence was lacking, especially after the realization that LECA already possessed mitochondria [12, 32]. Instead, they proposed that eukaryotes evolved from a symbiosis directly established between archaea and bacteria (Figure 2C).
The first detailed symbiogenetic scenario of eukaryogenesis was the Serial Endosymbiotic Theory proposed by Margulis [37, 53, 54]. She did not only propose that mitochondria and plastids derived from ancient endosymbiotic bacteria, but that the host that acquired those endosymbionts also evolved from a symbiotic event. The latter would have involved a wall-less, Thermoplasma-like, archaeon and spirochetes, which would have originally established a symbiosis by oxidising sulphide to sulphur required by the host [55] before evolving into eukaryotic flagella [37, 53, 54]. By contrast, Margulis thought that the nucleus, which Mereschkowsky believed of endosymbiotic origin as well [35], evolved autogenously [56]. Nonetheless, structural and phylogenetic evidence pointing to a symbiotic origin of eukaryotic flagella was missing, and apart from the idea of a symbiotic origin for the eukaryotic cell, Margulis’ hypothesis never became widely accepted.
In the decade of the 1990s, as knowledge of the biochemistry and molecular biology of archaea progressed, several models postulating a symbiotic origin of eukaryotes involving archaea and bacteria were proposed independently. Some of them did not put forward any particular mechanism, but relied on increasing evidence showing a chimeric nature of eukaryotes [57, 58]. Indeed, while eukaryotic informational processes are archaeal-like, the cell bioenergetics (the so-called “operational” genes) [30] and also the membrane phospholipids are bacterial-like [58, 59]. A few models proposed that metabolic symbiosis, or syntrophy, was the basis of the eukaryogenetic symbiosis [60]. One of them was put forward by D. Searcy, who suggested that eukaryotes derived from a sulphur-dependent Thermoplasma-like archaeon that would have incorporated a facultative anaerobic alphaproteobacterium able to oxidise sulphide to sulphur [61]. Two other models were more detailed: the Syntrophy and the Hydrogen hypotheses.
The Hydrogen hypothesis, by Martin and Müller, postulated that eukaryotes evolved from a methanogenic archaeon and an endosymbiotic and facultatively anaerobic alphaproteobacterium that would ferment organics in anoxic conditions (required for methanogenesis), liberating hydrogen that would serve for the reduction of CO2 to CH4 by the archaeal host [62]. The Syntrophy hypothesis, which we first proposed in 1998 [63] and refined in 2006 [64], invoked three symbiotic partners. First, a symbiosis based on interspecies hydrogen transfer established between an endosymbiotic methanogenic archaeon within a complex, myxobacterial-like, fermentative deltaproteobacterium, the host. A second endosymbiosis involving a metabolically flexible alphaproteobacterium, facultatively aerobic but also methanotrophic, would allow recycling methane within the symbiotic consortium and led to the evolution of the mitochondrion. This model had strong roots in microbial ecology, being based on widespread microbial interactions in anoxic settings [65]. Searcy’s model and the Hydrogen hypothesis converged in the proposal that the mitochondrial endosymbiosis and the origin of the eukaryotic cell were one and the same. The Hydrogen and Syntrophy models converged in proposing a similar metabolic basis (interspecies hydrogen transfer) for the eukaryogenetic symbiosis, but clearly differed in the number of symbiotic partners, the specific mechanism of eukaryogenesis and the selective forces put forward for the evolution of the eukaryotic nucleus.
Despite increasing support from phylogenomic data for the existence of only two primary domains, bacteria and archaea, instead of three [66, 67], which indirectly supported symbiogenetic models, these remained less popular than the mainstream, three-domain-based scenario whereby eukaryotes evolved from an independent proto-eukaryotic lineage sister to archaea. This prevalent idea changed radically with the discovery of Asgard archaea.
3. Shifting views on eukaryogenesis
3.1. Exploring the microbial world: Asgard archaea
The “molecular revolution” that started in the second half of the twentieth century did not only allow to infer universal phylogenetic trees but had additional, far-reaching implications for the study of microbial diversity, function and evolution in natural ecosystems. Although only a tiny fraction of microorganisms in the wild can be successfully cultured and studied in the laboratory, the study of genes and genomes directly from environmental samples has become fundamental to access information about this microbial “dark matter”. For many years, 16S and 18S rRNA gene amplicon sequencing has allowed the characterisation of prokaryotic and eukaryotic diversity in natural microbial communities, leading to the establishment of large curated reference databases containing many thousand sequences [68, 69]. While extremely useful to explore community composition over space and time, single-gene metabarcoding studies do not allow functional inferences based on protein-coding genes. For this, the development of high-throughput sequencing techniques and bioinformatic tools, with the possibility to generate metagenomes and metagenome-assembled genomes (MAGs), has become widespread and, together with single-cell genomics, is allowing access to the complete genome content of many newly identified microbial lineages [8, 70, 71, 72].
In 2015, Spang and co-workers published an article describing MAGs of a lineage of uncultured archaea, the Lokiarchaeota, assembled from deep-sea sediments in the vicinity of the Loki’s castle hydrothermal field in the North Mid-Atlantic Ridge [73]. This group of archaea had previously been identified by 16S rRNA gene amplicon sequences as Deep Sea Archaeal Group (DSAG) or marine benthic group B [74, 75, 76], but its metabolic potential and other genomic traits were unknown. Lokiarchaeota MAGs revealed the presence of a relatively high number of genes that had homologs exclusively in eukaryotes and were only sparsely found in other archaea. In addition, phylogenomic trees including eukaryotes placed them as sister to Lokiarchaeota, well nested within the archaeal tree [73]. This immediately raised interest in this group of archaea and soon after, members of the same research team released additional archaeal MAGs that defined several new groups forming a clade with Lokiarchaeota: Heimdallarchaeota, Thorarchaeota and Odinarchaeota. They were collectively called Asgard archaea, in reference to the pantheon of Norse gods [77]. These MAGs revealed the occurrence of even more of the so-called “eukaryotic signature proteins” (ESPs) shared with Asgard archaea, notably with the Heimdallarchaeota, to which eukaryotes seemed more closely related in phylogenomic trees [77] (Figure 3).
Schematic tree of archaea showing the currently most likely placement of eukaryotes based on phylogenomic analysis of highly conserved genes. Asgard archaea are classified in classes according to the GTDB taxonomy [78]. The DPANN cluster, originally named after its first recognized member lineages (Diapherotrites, Parvarchaeota, Aenigmarchaeota, Nanoarchaeota and Nanohaloarchaeota) comprises mostly reduced, parasitic members [8, 70, 71, 72]. TACK archaea (now classified in GTDB as the phylum Thermoproteota) include the original taxa Thaumarchaeota (GTDB Nitrosospheria), Aigarchaeota (GTDB Caldarchaeales), Crenarchaeota (GTDB Thermoprotei) and Korarchaeota (GTDB Korarchaeia) together with relative lineages (Bathyarcheia, Methanomethylicia). Euryarchaeota, one of the two archaeal clades defined by Woese together with Crenarchaeota [25, 26], is now divided in five separate phyla in GTDB (Halobacteriota, Thermoplasmatota, Methanobacteriota, Hadarchaeota, Hydrothermarchaeota).
Many more Asgard archaeal MAGs have been assembled to date. Most of them come from metagenomes of oxygen-deprived biotopes, including marine and freshwater sediments, microbial mats or hot springs [77, 79], although a few have been assembled from microoxic hypersaline lakes [80]. This has led to the description of the metabolic potential of many more Asgard archaeal clades, such as the Helarchaeota, potentially involved in the anaerobic oxidation of hydrocarbons [81]; the Hermodarchaeota, likely degrading alkanes and aromatic compounds coupled to nitrate reduction [82]; the Gerdarchaeota, using both organic and inorganic carbon [83]; the Sifararchaeota, possibly degrading polysaccharides and performing anaerobic methylotrophy [84]; the Freyrarchaeota and Wukongarchaeota, possibly carrying out homoacetogenesis [85]. Many of those archaeal groups have now been reclassified as classes or orders in the Genome Taxonomy Database (GTDB) [78], eukaryotes still placing deeply within or as sister to the Heimdallarchaeota in phylogenomic trees (Figure 3). The metabolic capabilities displayed by the different Asgard archaeal groups, as well as those inferred by ancestral metabolic reconstruction for their common ancestor, suggest that most of these archaea (and their ancestor) are strict anaerobes able to produce and/or consume hydrogen (electrons). This implies that these archaea are mostly involved in metabolic symbioses with other microorganisms that behave as hydrogen (electron) donors and/or scavengers [85].
The idea that Asgard archaea are involved in syntrophic interactions in nature seemed confirmed by the cultivation of the first two Asgard archaeal members, both belonging to the Lokiarchaeota. The first was Candidatus Prometheoarchaeum syntrophicum, which lives in syntrophy with either a sulphate-reducing deltaproteobacterium, a methanogen or both [86]. These partners serve as sinks for the hydrogen produced by the Asgard archaeon. The second cultured Asgard archaeon, Candidatus Lokiarchaeum ossiferum, also relies on syntrophy with a sulphate-reducer of the genus Halodesulfovibrio and a Methanogenium sp. methanogen [87]. Interestingly, both lokiarchaea exhibit a peculiar morphology with more or less long thin protrusions that can sometimes intertwine [86, 87]. They also reveal the presence of a complex actin-based cytoskeleton, as was predicted from MAGs [88].
The exploration of almost two hundred Asgard archaeal MAGs led to the identification of over 500 ESPs [85]. Most of them are involved in intracellular trafficking, secretion and vesicular transport. They include many GTPases, ubiquitin and ESCRT-III proteins, which are typically involved in membrane remodelling and vesicular transport in eukaryotes. This suggests that the eukaryotic ubiquitin-coupled ESCRT system has evolved through gene duplication and diversification from Asgard archaeal ancestors [89]. They also include actins and proteins, such as profilins [90] and gelsolin/cofilins [91], involved in the formation and dynamics of the actin cytoskeleton. Furthermore, the archaeal homologs of these proteins can complement eukaryotic proteins [90, 91], indicating that these archaea have a complex actin cytoskeleton and suggesting that the eukaryotic cytoskeleton evolved from its archaeal counterpart. Similarly, Asgard archaeal homologs of tubulin are much more closely related to eukaryotic tubulins than FtsZ, a prokaryotic homolog of the same gene superfamily involved in the formation of the cell division ring [92]. Furthermore, Asgard archaeal tubulins form protomers and protofilaments most similar to eukaryotic microtubules, although they assemble into ring systems more similar to FtsZ, constituting some sort of functional intermediate [93].
3.2. Eukaryogenesis by symbiogenesis
The discovery of Asgard archaea, which share many more genes with eukaryotes than other archaea, together with the phylogenetic position of eukaryotes within Asgard archaea, based on highly conserved genes, strongly support that eukaryotes evolved by some sort of symbiotic merging involving at least two prokaryotic partners, one Asgard-like archaeon and the alphaproteobacterial ancestor of mitochondria [73, 77, 94, 95, 96] (Figure 2C). Currently available data indeed support the existence of only two primary domains, Archaea and Bacteria [97]. Eukaryotes thus constitute a third, but secondary, domain of life.
Although microbial symbioses are plentiful in nature, particularly in oxygen-deprived environments, eukaryogenesis only happened once, suggesting that the mechanism that led to the stable integration of the symbiotic partners evolving into a complex cell was not that easy and/or involved contingent aspects difficult to reproduce de novo. Such a process likely required a more or less long co-evolution period during which many of the typical eukaryotic traits developed [38]. A first step would have been to yield the symbiosis obligatory, which might have happened by the transfer of one or more essential genes from the bacterial partner(s) to the archaeon followed by gene loss in the bacterial donor. From this step on, the consortium evolved as a single selective unit and the archaeal genome progressively became the future nuclear genome, gradually integrating more bacterial genes. Horizontal gene transfer between endosymbiotic and host genomes may have occurred by different means, from natural transformation to the mediation of viruses, transposons and other mobile genetic elements. Many of those genes coded for redundant functions in the archaeon. Accordingly, many of them were lost, but others were kept and subfunctionalised, giving rise to new functions [38]. Gene duplication and subfunctionalisation seem to have been at the origin of many gene families in eukaryotes [98, 99]. Some duplicated genes, being free from original selective constraints, evolved far beyond homology recognition, while other genes likely evolved de novo. The mix of archaeal and bacterial genes and the acceleration of evolutionary rate of many of them, readily explain the chimeric origin of eukaryotic genomes as well as the occurrence of specific eukaryotic genes.
In addition to genome evolution, the evolution of some typical eukaryotic traits such as the cytoskeleton and most of the membrane-remodelling systems, can mostly be traced back to the archaeal partner. Others, such as the operational functions related to the mitochondrion or membrane phospholipids, can be traced back to bacterial ancestors [38, 94, 100]. In summary, eukaryogenesis occurred by symbiogenesis, that is the physical merging of prokaryotic partners along a co-evolutionary process involving both chimerism and the formation of de novo traits. The latter was partly facilitated by the increased evolutionary rate of many duplicated genes and/or the evolution of truly novel eukaryotic specific mechanisms.
3.3. Current models of eukaryogenesis
Although current models of eukaryogenesis favour a symbiosis between an Asgard-like archaeon and at least one alphaproteobacterium at the origin of the eukaryotic cell, they vary on the mechanisms (the how) and the selective forces (the why) involved in the process. Based on the inferred type of symbiosis established by Asgard archaea with other microbes in nature, syntrophic interactions seem the most likely basis for the initial eukaryogenetic symbiosis [85, 96, 101, 102]. Currently, four different models propose a more or less detailed evolutionary scenario of eukaryogenesis based on syntrophic interactions. Other models suggest a more or less developed scenario, but do not point to any particular basis for the original symbiosis between the Asgard archaeon and its bacterial partner(s) (Figure 4).
Current models of eukaryogenesis based on the symbiotic merging of archaeal and bacterial partners. (A–C) Depict a selection of scenarios lacking a specified basis for the symbiosis. (D–E) Correspond to more detailed models postulating specific syntrophic interactions. (A) Phagocytosing Archaeon model: an archaeon develops cytoskeleton, endomembranes and phagocytosis prior to the engulfment of the mitochondrial ancestor [103]. (B) Inside-Out model: an archaeon develops extrusions that progressively engulf the mitochondrial ancestor [104]. (C) Pre-mitochondrial symbioses model: several bacterial symbiotic partners contribute genes and traits to an archaeon evolving into a proto-eukaryote prior to the mitochondrial symbiosis [100, 105]. (D) Hydrogen hypothesis: a fermenting alphaproteobacterium liberating hydrogen becomes an endosymbiont in an Asgard-like archaeon and triggers eukaryogenesis [106]. (E) Reverse Flow model: a complex hydrogen-producing archaeon becomes phagotrophic and engulfs its interacting alphaproteobacterial partner [101]. (F) Syntrophy hypothesis: a hydrogen-producing Asgard-like archaeon becomes an endosymbiont in a hydrogen-consuming sulphate-reducing deltaproteobacterium; mitochondria derive from a facultatively aerobic, sulphide-oxidizing alphaproteobacterium that also becomes endosymbiotic in the consortium [107]. (G) E3 (Entangle-Engulf-Endogenize) model: a hydrogen-producing Asgard archaeon in symbiosis with a sulphate-reducing deltaproteobacterium changes this partner by the facultatively aerobic mitochondrial ancestor [86].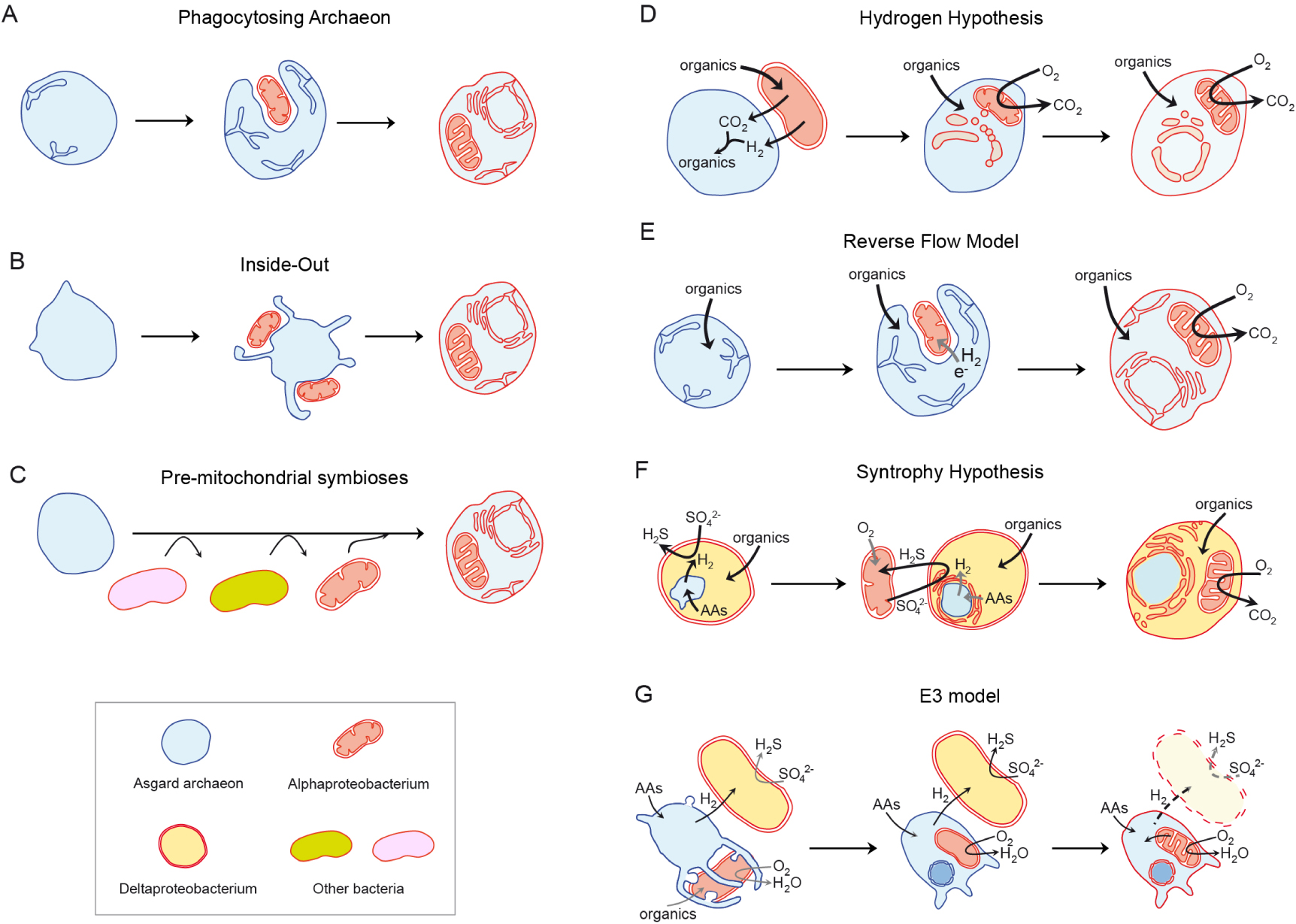
Another model that does not explicitly invoke a specific basis for the symbiotic interactions at the origin of the eukaryotic cell is the Pre-mitochondrial Symbioses (or Serial Symbiosis) hypothesis [100, 105] (Figure 4C). This model argues that a binary symbiosis between an archaeal host and the alphaproteobacterial ancestor of mitochondria does not explain the complex chimeric nature inferred for the eukaryotic ancestor. In particular, most of the genes of prokaryotic ancestry inferred to be present in LECA are of bacterial rather than archaeal origin, and from those only a relatively minor fraction is of clear alphaproteobacterial (or unassigned proteobacterial) origin. The remainder of those genes affiliate instead to Deltaproteobacteria, Actinobacteria, Bacteroidetes or Firmicutes, among others (Figure 5). In addition, genes of specific bacterial origin seem to correlate with coherent functions, which would be consistent with particular types of functions having been transferred to the archaeal host from privileged, long-term pre-mitochondrial symbiotic partners [100]. Accordingly, this model postulates that successive symbiotic interactions with other bacteria predated, and facilitated, the mitochondrial endosymbiosis that finally crystallised in the origin of the eukaryotic cell [105].
Phylogenetic origins of prokaryotic genes inferred to have been present in LECA. Numbers and relative proportions are based on Ref. [100, 105].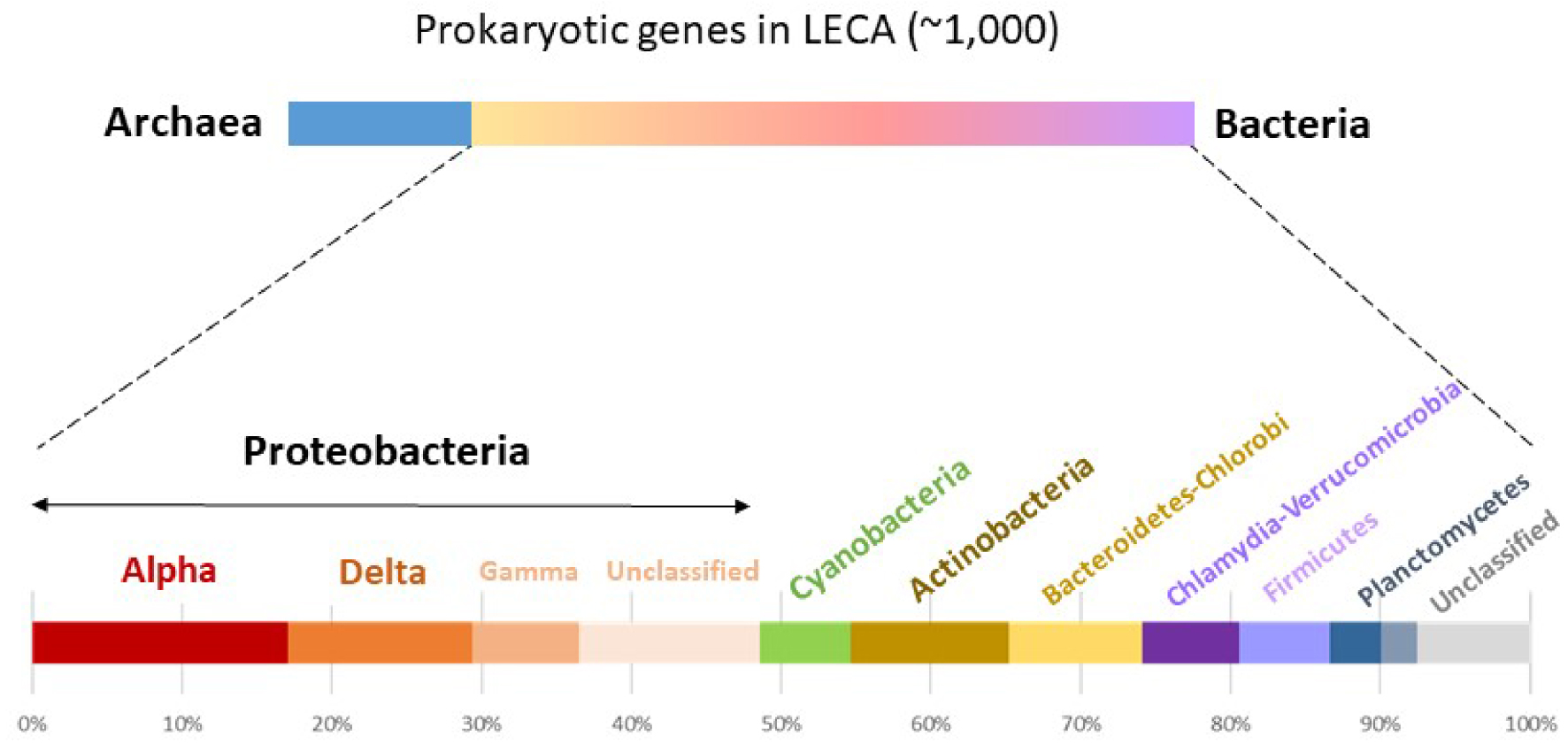
Among the current syntrophic models of eukaryogenesis, we find revised versions of the Hydrogen and Syntrophy hypotheses that now accommodate an Asgard archaeal partner. The revised Hydrogen hypothesis postulates that the incorporation of a hydrogen-producing alphaproteobacterium within a hydrogen-dependent autotrophic archaeal host triggered eukaryogenesis [106] (Figure 4D). Conversely, the Reverse Flow model, which is a syntrophy-adapted Phagocytosing Archaeon model, invokes a hydrogen-producing Asgard archaeon that progressively develops an endomembrane system and phagocytosis, which would subsequently allow to engulf a hydrogen (or electron)-consuming alphaproteobacterium (future mitochondrion) [101] (Figure 4E).
The revised Syntrophy hypothesis (or HS Syntrophy model) postulates that eukaryotes evolved from the initial incorporation of a hydrogen-producing Asgard archaeon (future nucleus) within a sulphate-reducing deltaproteobacterial host before the acquisition of the facultatively aerobic, sulphide-oxidizing alphaproteobacterial ancestor of mitochondria (Figure 4G). This is arguably one of the most detailed models in terms of the ecological context of the symbiosis and the evolution of metabolism, genome and endomembrane system, including the nucleus [107]. The Entangle-Engulf-Endogenize (E3) model proposed by the team that maintained the first Asgard archaeon in culture [86], posits that eukaryotes evolved from the progressive internalisation of an alphaproteobacterium (future mitochondrion), potentially a parasite, inside a hydrogen-producing Asgard archaeon. Based on the actual syntrophic interactions of Ca. Prometheoarchaeum, an initial tripartite symbiosis of those two partners with a sulphate-reducing deltaproteobacterium is proposed, but the latter would completely disappear from the consortium in subsequent steps (Figure 4F). In this sense, although three original partners are involved, only two, an archaeal host and the mitochondrial ancestor, would evolve into the eukaryotic cell, like most of the other models suggest.
Other models, often variants of the above, are being proposed. For instance, a more recent model proposes the intervention of an additional symbiont at the origin of the mitochondrial symbiosis. A second bacterium would have cooperated with the mitochondrial ancestor to kill prey with toxins and reactive oxygen species, before evolving into a digestive symbiosis through the acidification of protophagocytic cavities by respiration-derived proton export [108]. However, most scenarios of eukaryogenesis are partial and do not account for all the aspects involved in the evolution of the eukaryotic cell, leaving many open questions.
4. Open questions
Some questions on the origin of the eukaryotic cell, such as when and where eukaryotes evolved, or which specific lineages of Asgard archaea and Alphaproteobacteria were involved in eukaryogenesis, have partial answers or answers that will be refined with additional data and the improvement of phylogenomic and molecular dating analyses. The answer to other questions, such as how many partners participated in the original symbiosis or how the eukaryotic endomembranes and the nucleus evolved, will refute some models and potentially help to refine the remaining ones. In the following, we briefly evoke some of the current interrogations about eukaryogenesis.
Which and how many bacterial partners? Knowing which specific lineages of Asgard archaea and Alphaproteobacteria are closest to the respective prokaryotic partners involved in eukaryogenesis will possibly provide some additional clues about the metabolic potential of the original symbiotic consortium and perhaps help unravel the evolution of particular eukaryotic traits. The exploration of microbial diversity in natural ecosystems enriched in Asgard archaea might help identify new lineages potentially more closely related to those prokaryotic ancestors. Determining whether additional bacterial partners were involved in eukaryogenesis is especially crucial. In particular, several models involve more or less explicitly members of the Deltaproteobacteria [86, 100], although the Syntrophy Hypothesis makes the strongest case for their participation in the process [107]. Potentially, if some deltaproteobacterial members are found to harbor more genes/traits in common with eukaryotes, this can be indicative evidence for their implication in eukaryogenesis. For instance, some eukaryotic pathways can be traced back to myxobacteria (Deltaproteobacteria), including mitochondrial fatty acid beta-oxidation [109] and, most remarkably, given that membrane sterols are often considered hallmarks of eukaryotic membranes, steroid biosynthesis [110]. Nonetheless, it will be difficult to ascertain whether those pathways were acquired by horizontal gene transfer (HGT) by the Asgard archaeal partner (or by an intermediate pre-LECA symbiotic stage from co-existing bacteria in the environment), or by what is often called endosymbiotic gene transfer (EGT) from obligate symbionts taking part in the eukaryogenetic consortium. Additionally, intermediate situations are possible, with two or more bacterial symbiotic partners participating to eukaryogenesis, but with HGT from external bacterial donors to LECA or pre-LECA stages.
When and where did eukaryotes evolve? Current data based on microfossils that are unambiguously interpreted as eukaryotic, with traits indicative of a developed cytoskeleton and external decoration, support the idea that eukaryotes had already evolved by the mid Proterozoic (1.5–1.6 Ga) [111, 112]. These dates are consistent with those obtained by some molecular dating analyses, which suggest that LECA lived in the 1.0–1.6 Ga time frame [113]. As the microfossil record, but also the genome sampling across the eukaryotic tree and the molecular dating methods, improve, narrowing these dates might be foreseeable. However, it might be more difficult to point out the date at which eukaryogenesis started. A likely older boundary seems that of the evolution of oxygenic photosynthesis and the start of the oxygenation of the atmosphere, which happened at around 2.4–2.5 Ga, marking the beginning of the Great Oxidation Event (GOE) [107, 114]. Oxygen availability would have fostered the evolution of aerobic respiration, dangerous because of the resulting oxidative damage, but also much more efficient in terms of energy yield than anaerobic respirations. The ancestor of mitochondria was likely a facultative aerobe, respiring oxygen but being able to ferment and to respire fumarate or nitrate. It probably lived in microoxic environments and might have been able to move along the redox gradient established in sediments or microbial mats. This would have facilitated its interaction with Asgard archaea [107], which are mostly and ancestrally strict anaerobes located in deep sediment layers [77, 85].
Was phagotrophy a requirement for eukaryogenesis? This question is tightly linked to that of whether mitochondria evolved early or late, which is often considered as discriminant for eukaryogenetic models. For the Hydrogen hypothesis, eukaryotic-like phagotrophy was not strictly needed to incorporate the alphaproteobacterial endosymbiont into the Asgard archaeon, and this initial endosymbiosis was the cause triggering eukaryogenesis [62, 106]. By contrast, for the Phagocytosing Archaeon and related models [101, 103], the evolution of an endomembrane system and phagocytosis was mandatory prior to any possible bacterial engulfment; in a sense, an already engaged eukaryogenetic process was required for the mitochondrial ancestor to be engulfed. These latter models relied on the idea that prokaryotes lack a complex cytoskeleton and endomembrane systems. However, many prokaryotes do harbour endomembrane systems, including cyanobacterial thylakoids and membranous compartments in proteobacterial methylotrophs, magnetotactic bacteria or anammox planctomycetes [115, 116, 117]. Some bacteria, such as the planctomycete Gemmata obscuriglobus [118] and members of the phylum Atribacteria [119], even harbour nuclear-like compartments. Endomembranes in archaea have seldom been described, but seem to exist as well [120]. Likewise, bacterial endosymbionts exist within other bacteria, despite the internalization mechanism being unknown [121]. Finally, some planctomycete bacteria can carry out phagocytosis via a novel mechanism [122]. These observations suggest that prokaryotes, notably bacteria, can develop endomembranes and engulf other bacteria without the need for eukaryotic-like phagocytosis. Thus, from a mechanistic point of view, an ongoing eukaryogenetic process does not seem a requirement for the incorporation of endosymbiotic prokaryotes.
How and why did the nucleus evolve? Although the nucleus is arguably the most idiosyncratic feature of the eukaryotic cell, few models of eukaryogenesis actually address its origin. The nuclear membrane is continuous with the endoplasmic reticulum and, from a mechanistic point of view, it seems reasonable to think that both evolved together by invagination of the plasma membrane thanks to membrane-remodelling systems of mostly archaeal origin (ESCRT, actin cytoskeleton, GTPases) [89, 123, 124, 125, 126]. However, the evolution of more specific features, such as the nuclear pore, remains more enigmatic [94]. Certain models even speculated that the eukaryotic nucleus originated from viruses [127, 128], based notably on the putative resemblance of the so-called “viral factories” (subcellular locations where viral replication and morphogenesis take place in some eukaryotes) to nuclei [129]. Giant viruses were favourite candidates for a hypothetical viral eukaryogenesis, as they contain several homologs of typical cellular genes and were even proposed to form a fourth domain of life, sister to eukaryotes [130, 131]. However, by contrast with cells and their membrane systems, which divide from pre-existing ones (omnia cellula e cellula), viruses and cells or cell nuclei lack any structural continuity. Viruses are assembled de novo at each generation, and mechanisms explaining a putative evolutionary and physical continuity between viral factories and cell nuclei are lacking. Furthermore, phylogenetic analyses using complex sequence evolution models (allowing to cope with strong differences in evolutionary rate) and a comprehensive taxon sampling clearly showed that cell-like genes in giant viruses were transferred to the viral genomes from their hosts, and consequently do not reflect a viral ancestry of eukaryotes [132, 133, 134, 135, 136].
The selective forces that led to the evolution of the nucleus remain mysterious [94]. Some authors suggested that the nuclear compartment evolved to prevent chromosome shearing by cytoskeletal movement during mitosis [137, 138]. However, many eukaryotes disintegrate the nuclear membrane during mitosis (open mitosis) and cope well with cytoskeletal movements; also, eukaryotes have several or many linear chromosomes and powerful DNA repair mechanisms, which should limit any potential mechanical breakage induced by cytoskeletal movement [94]. In the case of the Syntrophy hypothesis, the nucleus had a partial autogenous origin. The genome was of archaeal origin, deriving from the archaeal endosymbiont, but the nuclear membrane evolved from a bacterial secretory endomembrane system that developed to transport hydrolytic enzymes, whose synthesis became progressively centralised by the archaeon, to the bacterial periplasm. The nuclear membrane promoted the uncoupling of transcription and translation, facilitating intron spreading, and was subsequently retained to prevent aberrant protein synthesis [64, 107]. Intron spreading was claimed to be the major trigger for the evolution of the nuclear membrane within the framework of the Hydrogen hypothesis; the nuclear membrane would have evolved to uncouple transcription and translation, thus avoiding the deleterious synthesis of aberrant proteins [139]. However, as soon as an intron interrupts an essential gene in the absence of an efficient splicing mechanism and transcription–translation uncoupling, natural selection would eliminate these cells. Thus, in this case, a selective mechanism explaining intermediate steps in the evolution of the endomembrane system and the nuclear membrane needs to be put forward.
Bacterial-like eukaryotic membranes, what origin? Bacterial and archaeal phospholipids forming the plasma membrane are radically different in terms of their constituents and properties. Eukaryotic membrane phospholipids are of bacterial type. Archaeal phospholipids are composed of isoprenoid lateral chains linked by ether bonds to glycerol-1-phosphate [140, 141]. This kind of membrane phospholipids confer archaea a higher resistance to various extreme conditions; in particular, they prevent proton leakage at high temperature [59]. Bacterial phospholipids are composed of fatty acid moieties linked by ester bonds to glycerol-3-phosphate. It has been known for a long time that several bacteria can use ether linkages, notably thermophilic ones [142, 143], but also eukaryotes. Therefore, although the ether/ester linkage constitutes part of what is known as the “lipid divide” [144], the real divide between archaeal and bacterial phospholipids is determined by the opposite stereochemistry of the glycerol-phosphate moieties [141, 145], in combination with the nature of the lateral chains. Both archaea and bacteria synthesise fatty acids and isoprenoids, but these are exclusively incorporated by one or the other domain into their membrane phospholipids [146]. Except for the Syntrophy hypothesis, which proposes a bacterial host and, accordingly, bacterial origin of the plasma membrane and endomembranes (the archaeal membrane would be lost during the evolution of the nucleus [64, 107]), most models of eukaryogenesis postulate an archaeal host [86, 101, 104]. Thus, these models must invoke an archaeal-to-bacterial membrane phospholipid transition that is difficult to explain taking into account that the archaeal membrane (including its proteome, which was adapted to the local physicochemistry of archaeal lipids), was adapted to interact with the environment. It is difficult to see which selective force would promote such a radical change of membrane phospholipids and associated proteome [94]. Nonetheless, although archaeal–bacterial membrane transitions have never been observed in nature, liposomes of heterochiral (archaeal + bacterial) phospholipids seem to be stable [147]. Attempts to carry out a bacterial-to-archaeal membrane phospholipid transition have been made in Escherichia coli by heterologously overexpressing archaeal-phospholipid biosynthesis genes [148]. However, expression levels that result in the incorporation of more than 30% archaeal phospholipids in E. coli impair growth, cause anomalous cell division and secretion of vesicles, and are, in fine, deleterious [148]. Future experiments should show whether such a transition is possible. However, it remains unclear whether that transition was feasible in the natural microbial communities in which eukaryotes evolved, as the fitness of transition stages may have been too negatively impacted for the hypothesised transition to occur.
5. Perspectives
The exploration of the microbial diversity in poorly studied, oxygen-deprived, ecosystems has facilitated the discovery of the Asgard archaea, which share many genes with eukaryotes and are their closest relatives in phylogenomic trees based on highly conserved markers. This discovery has promoted a shift in the type of scenarios for the origin of the eukaryotic cell, which now favour some sort of metabolic symbiosis between archaea and bacteria as the starting point of eukaryogenesis. While existing models try to account for some aspects of how this evolutionary process took place, very few are detailed enough to explain the evolution of all typical eukaryotic features. More explicit models are needed, in particular to explain the origin of the nucleus, the bacterial-like membrane phospholipids of eukaryotes, and the bacterial heritage in eukaryotic genomes. Together with improved phylogenomic analyses and a better exploration of the microbial world, an increased understanding of the biology and the ecology of the prokaryotic lineages closest to eukaryotes should help to discriminate between existing models and/or to refine some of the existing ones. New eukaryogenesis models should propose plausible processes that account for observed traits and phylogenetic patterns in a realistic ecological context.
Conflicts of interest
Authors have no conflict of interest to declare.
Acknowledgments
This research was funded by the European Research Council Grants ProtistWorld (322669, PL-G), PlastEvol (787904, DM) and the Moore-Simons Project on the Origin of the Eukaryotic Cell, Moore Foundation grant GBMF9739 (PL-G; https://doi.org/10.37807/GBMF9739).




 CC-BY 4.0
CC-BY 4.0