1. Introduction
Fractals are intriguing by their mathematical and geometric beauty [1]. Their intricate shapes are made up by the repetition of a similar pattern at every scale. Despite their complexity, they can be easily simulated using intensive recursive computation on standard computers (see e.g. [2]). Biology provides a wealth of illustrations of fractal-like shapes. However, none of the computational algorithms used to generate fractals consider the complex chemical and physical processes responsible for the development of biological shapes, therefore giving little insight into their origins in nature, which thus remains largely unknown.
Cauliflower curds provide a unique opportunity to address this question. Their fractal-like shape derives from a modification of the flower development process. Strikingly, a cauliflower-like severe change of morphology is produced by mutations of the APETALA1 (AP1) and CAULIFLOWER (CAL) genes in Arabidopsis thaliana [3, 4], a brassicaceae from the same family as the cauliflower and broccoli (Figure 1a, b). In a recent work [5], we provided a mechanistic explanation of how these mutations alter flower development, by recursively triggering lateral meristem development, which results in the fractal-like shape of the curds. Moreover, our work showed how different cauliflower morphologies can be caused by changes in the regulation of plant growth dynamics. In the following, we explain the main results and contributions of our work. In the first section, we describe how some mutations of the gene regulatory network (GRN) involved in flower development can open the way to the development of iterative structures necessary to produce fractals. Then, we explain how this GRN regulates the growth dynamics of plant axes and induces a chain reaction of growing axes leading to the emergence of fractal curds. We then describe how a simple model of plant architecture development was used to show that the changes in plant growth dynamics can be at the origin of the remarkable Romanesco cauliflower. Finally, we briefly discuss the relevance of our findings in the context of plant domestication.
WT vs ap1 cal morphology. (a) WT plant with an inflorescence meristem producing flower meristems. (b) The cauliflower morphology produced by the overproduction of lateral meristems in the ap1 cal mutant. (c) A microscopy image of an inflorescence meristem (IM), surrounded by flower meristems (FM). Photographs credits: Jan Trass (c) and Marie Le Masson (a and b).
2. The gene regulatory network
Aerial plant meristems are structures composed of undifferentiated cells that are located at the most apical part of shoots. These meristems produce lateral meristems that can take different identities (e.g. leaf, inflorescence (IM) or flower meristems (FM)) depending on the regulatory genes they express (Figure 1c). The gene expression in meristems is regulated by interactions between master regulatory genes. In particular, in A. thaliana, shoot meristems express TERMINAL FLOWER1 (TFL1), a shoot identity gene that prevents meristems from differentiation by repressing flower identity genes, such as APETALA1 (AP1), CAULIFLOWER (CAL) and LEAFY (LFY) [6, 7, 8, 9, 10, 11] (Figure 2a). During the vegetative phase of plant development, flower identity genes are inactive and lateral meristems take shoot and leaf identity. However, as plant ages, flowering signals induce the expression of AP1, CAL and LFY in new lateral meristems (Figure 2b), which turns them into flowers. AP1 inhibits shoot identity by repressing TFL1 in FM [7, 12, 13] and stabilises the expression of floral regulators by forming a positive feedback loop with LFY (Figure 2a) [6, 7, 8, 9, 10, 14, 15, 16, 17]. In A. thaliana, the double mutant of ap1 cal, produces cauliflower-like structures instead of flowers [3, 4, 6]. Those structures correspond to meristems initiated as floral (with LFY expression) but that revert to shoot (with TFL1 expression) because the AP1/CAL-LFY positive feedback is missing (Figure 2c).
Gene regulatory network and gene expression profiles involved in the flowering transition. (a) Interactions between the major floral regulators. Gene expression during late development in the shoot and lateral meristems of wild-type (WT) (b) and ap1 cal mutant (c) plants. SAM = shoot apical meristem. FM = Floral meristem (lateral meristem). In green and red, TFL1 and LFY expression, respectively.
The first attempts to model the floral GRN only based on TFL1, AP1/CAL (AP1 and CAL were included as a single variable because they are homologous genes with redundant functions) and LFY, led us to realise that there were missing elements in the network. In particular, those inducing the expression of TFL1 were lacking. We therefore looked for possible candidate genes and identified additional genes intricately linked to the original master regulators. These genes are the MADS-box genes SUPPRESSOR OF OVEREXPRESSION OF CONSTANS1, AGAMOUS-LIKE24 and XAANTAL2 (collectively renamed the “SAX” genes). Interestingly, on the one hand, SAX proteins promote the expression of the flowering genes AP1 and LFY [14, 18, 19, 20] and repress TFL1 forming a complex with AP1 [11]. On the other hand, work from our collaborators showed that SAX proteins are also able to activate TFL1 expression. We updated the GRN with this information (Figure 3), formalised these interactions in a model of ordinary differential equations and analysed both its transitory and stationary properties. These analyses showed that the GRN was sufficient to produce the expected patterns of gene expression in a wild-type (WT) plant, in an ap1 cal mutant, and in the other single mutants of the different genes present in the network. In addition, the model predicted a high level of SAX expression in ap1 cal meristems that is not observed in WT plants and we experimentally confirmed this prediction with the SOC1 gene [5].
Gene regulatory network of the main genes involved in early floral development (Image from [5]. Arrows represent positive regulations, flat head arrows represent negative regulations and the merging arrows from SAX and AP1 towards TFL1 represent the formation of the SAX-AP1 complex. F and Auxin are inputs of the model. The equations of the model are available in [5].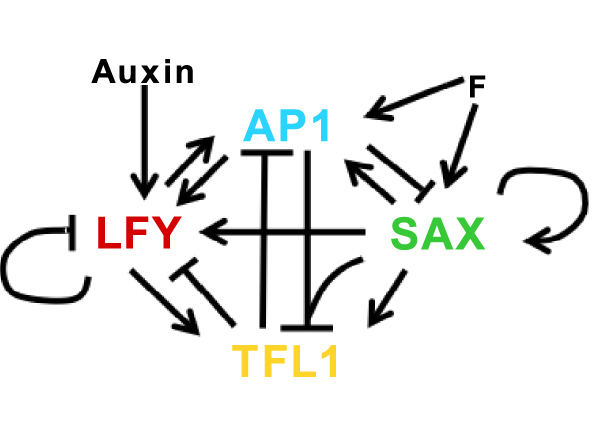
Having found a minimal GRN able to recapitulate the different gene expression states and explain the shoot identity of new lateral meristems in the ap1 cal mutant, we wondered whether it was sufficient to explain the cauliflower inflorescence morphology.
To address this question, we first took into account that the plant architecture does not solely depend on the fate of its meristems, but also on additional developmental factors such as phyllotaxis of the shoots, the growth rate of the internodes, the time it takes for newly created meristems to produce new lateral organs, etc. Moreover, we also noticed that the GRN final states for lfy and ap1 cal mutant simulations are identical with a high level of TFL1 and SAX and a low level of AP1/CAL and LFY, indicating that the architecture of the plant cannot be solely explained by the GRN stationary states. To understand the origin of curd morphology, we were thus led to analyse in greater depth the dynamics of the different actors in the growing branching structure. For this, we designed a 3D computational model simulating the growth of a plant whose development dynamics was regulated by the state of GRNs computed in each meristem. We call this model, the architectural model.
3. The architectural model
More precisely, our architectural model comprises two main components, meristems and internodes. Each meristem has its own identity based on the state of its GRN (Figure 4a). The identity of a meristem determines its behaviour. Vegetative and inflorescence meristems produce internodes and new lateral primordia (meristems) at each plastochron. Flower meristems turn into flowers, thus ending the axis growth. Vegetative meristems have low TFL1, AP1 and LFY expression. Inflorescence meristems have high TFL1 expression and low AP1 and LFY expression. Finally, flower meristems have low TFL1 expression and high AP1 and LFY expression. At each step, the model inputs a level of flower inducing signal (that was assumed to increase in each meristem with aging) and a level of auxin that was set maximal in each new meristem and then exponentially decay. The state of the GRN was thus reevaluated in each meristem at each time step and was used to determine the identity of the meristem. In addition, since most meristems of order 3 remained inactive in A. thaliana, we added a rule to the model, specifying that meristems of order 3 should be kept dormant, irrespective of their identity (Figure 4b).
The architectural plant model. (a) Schematic description of the architectural plant model. Green circles represent inflorescence/vegetative meristems. Red circles represent flower meristems. Red lines represent internodes. The simulation starts with a vegetative meristem. Vegetative and inflorescence meristems were able to produce lateral meristem and internode, therefore driving plant growth with time. Each meristem contains its own GRN. The state of the GRN determines its identity. (b) WT morphology produced by the model vs a photograph of a WT A. thaliana. The arrows indicate the meristems order. In (c) the left panel shows that the original model produced cauliflowers with meristems of maximal order 3. In the middle, a photograph showing an ap1 cal plant with a meristem order higher than 3. On the right panel, a simulation assuming that LFY expression releases the production of meristems of orders higher than 3.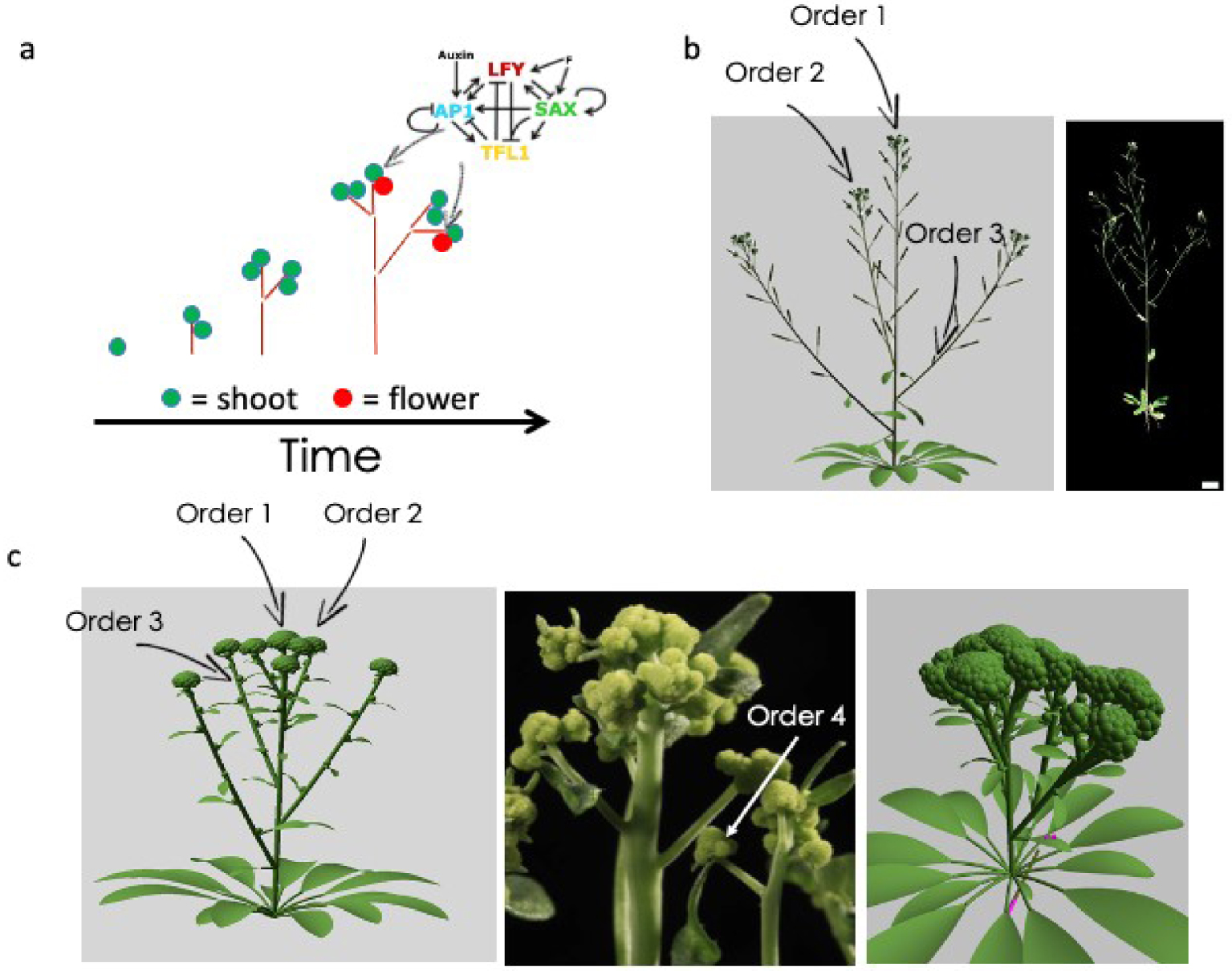
This model, calibrated for WT plant development, was able to recapitulate morphologies of mutated plants with the expected architecture (Figure 4b). For example, tfl1 mutants terminate with the production of a few flowers due to AP1 and LFY expression in the shoot apical meristem [21], lfy mutant produced mostly secondary branches [22]. However, the model could not reproduce the ap1 cal morphology. First the ap1 cal inflorescence, contrary to lfy, is mostly devoid of leaves and no element was accounting for this in the model. Second, the cauliflower phenotype arises from the accumulation of meristems on the top of each other, reaching orders higher than 3 while we had imposed a limitation to the growing orders to model the wild-type plant properly (Figure 4c).
We then realised that, despite the fact that the lfy mutant and the ap1 cal mutant have identical stationary states in the inflorescence lateral meristems, a critical difference between the two mutants lies in the dynamics to reach that state. While in the lfy mutant, LFY expression never rises up, a transient peak of LFY expression is produced in lateral meristems of ap1 cal mutants before they finally take a shoot identity. This key difference was enough to explain both previous issues. Indeed, it was already well documented that the presence of LFY is responsible for repressing bracts in primordia [22]. Second, previous work from our group [23] had shown that ectopic expression of LFY was able to allow high order meristems to grow. The transient rise of LFY in ap1 cal was thus essential to free meristem outgrowth at all orders and induce a chain reaction.
In conclusion, our work showed that the cauliflower phenotype is mainly due to the fact that in the ap1 cal mutant there is an unsuccessful flower development that produces undifferentiated meristems instead of flowers. The transient peak of LFY expression in the ap1 cal mutant is responsible for the activation of all meristems and causes the recursive production of new meristems at all orders. In other words, the fractal-like cauliflower morphology is the result of an alteration of two fundamental processes shaping plant architecture, namely meristem differentiation and meristem outgrowth repression.
4. The Romanesco hypothesis
Brassicas curds have different fractal morphologies. Cauliflowers exhibit a curd with a round dome. Their fractal nature can be observed with a curd morphology observable at many scales, each curd being formed of smaller curds with similar round domes. This fractal pattern is reinforced by the typical spiral arrangement of smaller curds forming each larger one. The fractal structure of the Romanesco in nested spiralling curds is similar, but the conic shape of curds dramatically highlights its fractal geometry (Figure 5a, b).
Cauliflower and Romanesco morphologies produced by the model and experimentally. (a) Cauliflower and (b) Romanesco morphologies. Curd morphologies produced by the model with (c) constant plastochron vs (d) accelerating plastochron. (e) ap1 cal curd morphology vs (f) clv3ap1 cal curd morphology. Photograph credits: Marie Le Masson.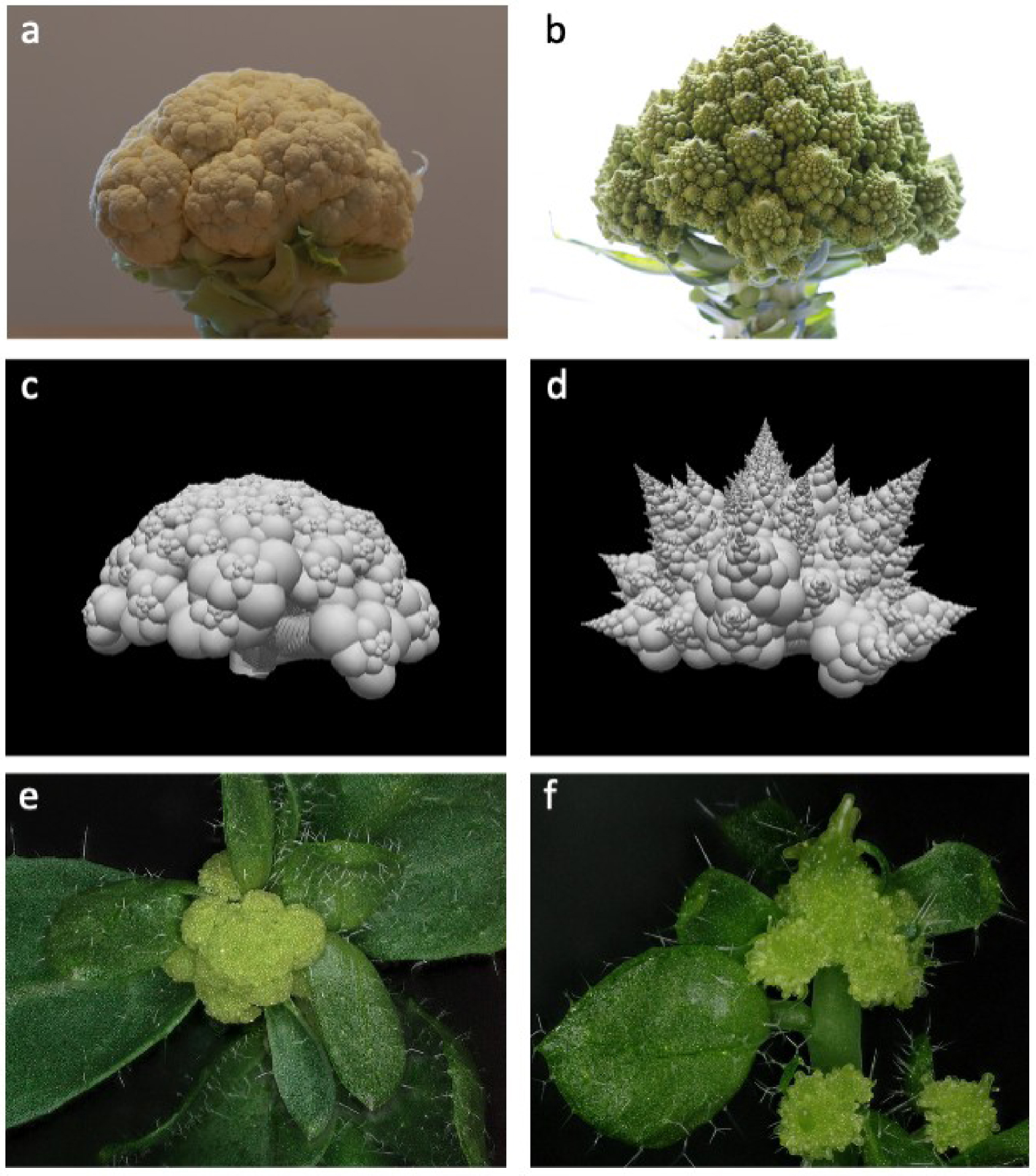
We thought of taking advantage of our architectural model to better understand what regulates the development of the different curd morphologies. A previous comparison between cauliflower and Romanesco curds identified several features that are constantly increasing during development in Romanesco curds, but that are constant in cauliflowers [24]. These properties are: (1) the production rate of new lateral meristems; (2) the number of phyllotaxis spirals in clockwise and counter-clockwise directions; (3) the number of lateral primordia produced by a meristem before the first one starts producing its own lateral primordial (meristem onset delay); and (4) the size of the meristems. Whether some of these parameters are causal to the Romanesco phenotype had remained elusive. However, we reasoned that according to phyllotaxis studies [25, 26, 27, 28], these properties must be linked to each other. Consequently, we wondered if the morphological curd variations could also be explained by the difference in any parameter controlling these properties. To check this, we constructed a simplified version of our architectural model, making it possible to test the consequences of changing these parameters on the final curd morphology. Strikingly, we found that an increase in the meristem production rate was sufficient to produce conspicuous Romanesco-like shape whereas constant values of this parameter produce a normal cauliflower morphology (Figure 5c, d). This acceleration in the meristem production rate induces the insertion of more internodes in the meristem’s axis. Assuming that this process does not affect the elongation law of the internodes (internode elongation was kept unchanged in the simple model), this acceleration of the meristem production rate indirectly induces an acceleration of overall stem elongation relative to its lateral axes, which results in conspicuous conical structures with very visible spirals themselves made of cones and so on.
According to our model, increasing the production rate of lateral meristems should transform a cauliflower into a Romanesco curd. To test this hypothesis in planta, we used the fact that, according to phyllotaxis models, meristem production rate may in principle be augmented by an increase in the size of the central zone. In A. thaliana, the clavata (clv) mutants (e.g. clv1 and clv3) are known to produce bigger meristems [29, 30]. Thus, we crossed a clv3 mutant with the ap1 cal double mutant. Indeed, the clv3 ap1 cal triple mutant has a modified curd shape, with a morphology being more pyramidal than for ap1 cal curds (Figure 5e, f). This result suggests that the meristem production rate is a fundamental regulator of the final curd morphology. Interestingly, CLV3 and a couple of other related genes seem to have a lower expression in Romanesco curds than in cauliflowers [5], but the causal link between this observation and the curd phenotype needs to be studied.
This mechanism provides a first evidence that the strikingly different morphologies observed in curds could be due to simple change in a single parameter, such as the size of the meristem central zone. This changes meristem production rate, which is then propagated throughout the whole plant as a change in the relative elongation rates of the different axes, resulting in a dramatic change of the curd global morphology.
Altogether, our results provide a plausible explanation for the appearance of fractal-like structures in plants and for the possibility to significantly modify their geometry based on variations of only a limited number of growth regulation factors.
5. Discussion
The pervasive presence of fractal-like structures in biology is intriguing. Until now, the developmental origin of most biological fractal-like structures was still unknown. However, deciphering their origins is a non-trivial task due to the high interconnectivity and non-linearity of developmental processes, which take place at multiple scales. In our work, we used modelling and experimental tools to study the origin of fractal-like cauliflower structures. Our results showed that the fractal structure of curds is very likely the consequence of three main mechanisms. First, the mutation in the ap1 cal genes produces an unsuccessful flowering transition. As a consequence, all meristems take a vegetative or inflorescence identity. The ap1 cal mutation also releases the production of lateral meristems at all orders. This is due to the transitory expression of LFY in the mutant. Finally, a Romanesco-like form of the curds can be induced when extra mutations induce an accelerated production of new lateral meristems. Our work provides a first plausible explanation of how fractal-like structures develop linking gene activity with macroscopic structures.
Our work also provides a framework to better understand fundamental aspects of plant development and plant domestication. The unexpected discovery that a transient exposure to LFY can have a profound effect on how the meristem responds to general cues such as those limiting the growth of high-order structures is not understood at the molecular level and it would be interesting to identify which LFY target genes exert this function.
Moreover, even if our work pointed towards flower and inflorescence regulators that can be responsible for the development of curds, the mutations that were selected during the long process of cabbage, broccoli, cauliflower and Romanesco domestication still need to be identified [31] and validated using genetic and functional analyses. Genome comparisons are a powerful tool, but yield many possible candidates that until now, except for mutations affecting the CAL gene, have not yet been validated [32]. We believe that our models can provide a very useful framework for their identification.




 CC-BY 4.0
CC-BY 4.0