1 Introduction
The discovery of the naturally-occurring, heavy-chain-only antibodies (HCAbs) in Camelidae [1,2] presents an alternative approach in pharmaceuticals and drug treatment [3]. These antibodies are easily expressed in bacteria and amenable to recombinant DNA technology. Derivatives of these antibodies are stable, versatile, and have unique refolding capacities, reduced aggregation tendencies, and high affinity to their cognate antigens. The antigen-binding site of these antibodies is confined to one domain referred to as the variable domain of the heavy-chain or VHH. These variable nanoantibodies or Nanobodies® (Nb) can be easily expressed in bacteria, yeasts or in other hosts as recombinant single-domain antibodies (sdAb), which are considered as the smallest available intact antigen-binding fragments [4]. Due to their biophysical and pharmaceutical properties, Nb fragments offer broad application potentials, especially as new immunotherapeutic drugs and also as efficient treatment and diagnostic methods [3].
The overall structure of mammalian immunoglobulin-γ (IgG) antibodies comprises two heavy (H) and two light (L) identical, highly conserved polypeptide chains [5]. In addition to the conventional heterotetrameric antibodies, sera of Camelidae also possess two other special types of IgG antibodies (Fig. 1a). These antibodies are devoid of the L chain polypeptide and composed of three instead of four globular domains as they lack the first constant domain (CH1). The H chain of the homodimeric protein at the N terminal domain contains the variable domain VHH, which serves to associate it with its cognate antigen [4]. This domain is adapted to become functional in antigen-binding in the absence of variable-light (VL) chain domain [6–9].
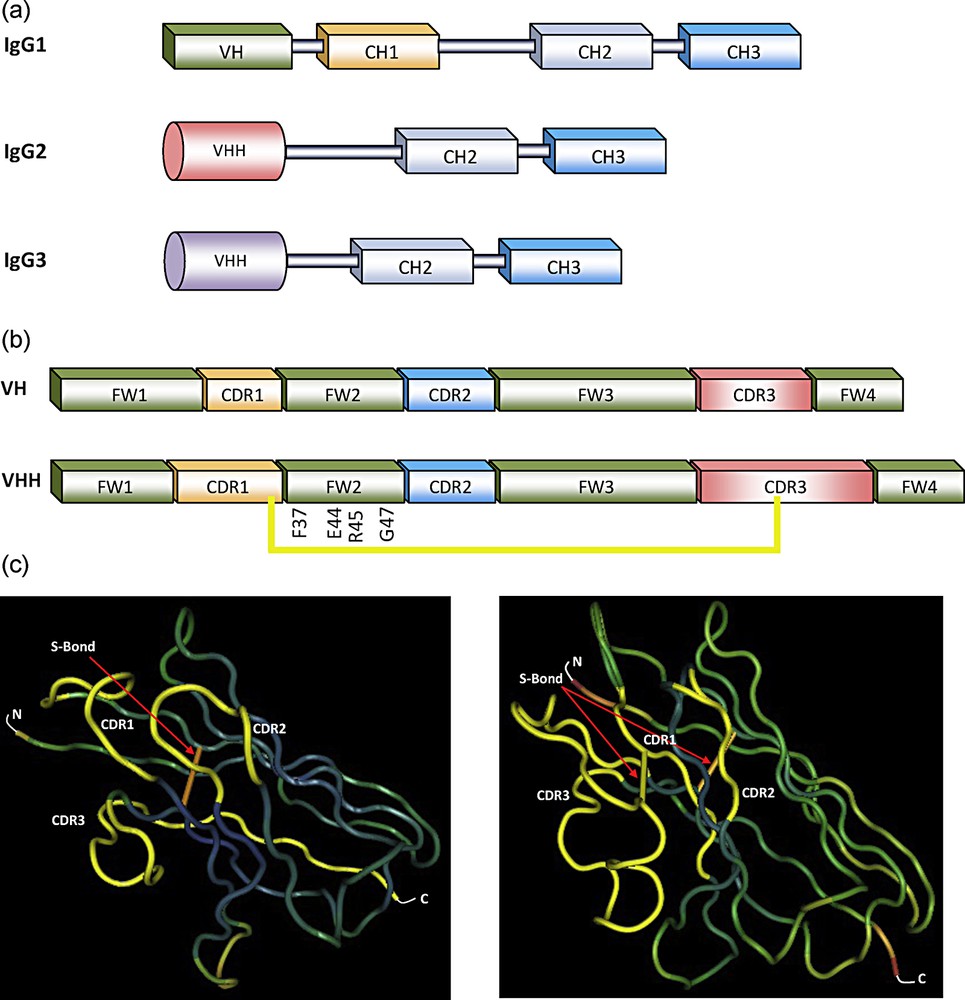
(a) Schematic representation of antibodies in camelids sera [29]. IgG1: conventional heavy-chain antibody (HCAbs) (VH, CH1, hinge, CH2 and CH3), IgG2 and IgG3: two types of homodimeric heavy-chain antibodies containing only H chains (VHH, hinge, CH2 and CH3). The last two IgGs differ in the length of hinge. (b) Sequence organization of the VH and VHH with four frameworks (FW) and three CDRs. The hallmark amino acids in FW2 within the VHH are given, as well as the additional disulfide (S) bond between CDR1 and CDR3 (yellow). (c) 3-D structure of the VH and VHH domains, showing the two disulfide (S) bonds between FW1 and FW3 (orange) and between CDR1 and CDR3 (yellow). For interpretation of references to color, see the online version of this article.
The organizations between the variable domain of the H chain of classical antibodies (VH) and that of HCAbs (VHH) are remarkably similar as both are composed of four conserved sequences, namely the framework regions (FWs), surrounding three hypervariable regions, namely the complementarity determining regions (CDRs) (Fig. 1b). However, VH and VHH have important differences [10]. Based on amino acid sequences, it was evident that the sizes of CDR1 and CDR3 of VHH are larger than those of VH. FW region in VH domain contains more hydrophobic amino acids, while VHH domain contains more hydrophilic amino acids (Fig. 1b). This distinction explains the absence of VL association and the solubility of VHH as a single-domain entity [11]. In addition, VHH contains an additional disulfide bond (Cys–Cys) [10] that assists in shaping the loop structure [12,13] (Fig. 1b and c).
Most VHHs recovered to date have been isolated from immunized camels [14–17]. The procedure is time-consuming, costly and likely redundant for certain antigens, hence, naïve VHH libraries have been proposed [15]. The rationale of using this type of library is that it is more diverse, which allows one to identify binders for any potential antigen [14,18].
In this work, we describe the construction of a naïve library from camelid sera and show that it allows recovery of a more diverse library suitable for the standard immune applications and amenable to recombinant DNA technology. Its production depends on bacteria; hence, it is significantly less expensive than mammalian cell cultures.
2 Materials and methods
2.1 Blood collection and RNA purification
Twenty-milliliter blood samples were collected from 30 young non-immunized camelids in EDTA, and lymphocytes were isolated using Ficol separation method [19]. About 109 cells were collected and divided into 2-ml microfuge tubes; RNA purification was performed using QIAamp® RNA blood extraction mini kit (Qiagen, Düsseldorf, Germany) according to the manufacturer's instructions. Then, 2 ug RNA were used in cDNA synthesis using RevertAid reverse transcriptase (cat. No. EP0441, thermo scientific, KMG Biotech, RAK, United Arab Emirates) and polyT-18 primer.
2.2 VHH amplification and library construction
Stepwise PCR amplification (Fig. 2) was done to recover the highly variable vhh gene for library construction. Ten ul cDNA were used for PCR 1 of vhh gene using a forward primer (VHBACKA6: 5′-GAT GTG CAG CTG CAG GCG TCT GGR GGA GG-3′), with PstI recognition site (underlined) indigenously available at the 5′ end of the gene, and reverse primer (CH2FORTA4: 5′-CGC CAT CAA GGT ACC AGT TGA-3′) [20]. PCR conditions were: 1 cycle at 94 °C for 2 min, 25 cycles at 94 °C for 30 s, 52 °C for 60 s, 72 °C for 60 s and finally 1 cycle of 72 °C for 10 min. Amplicons ranging from 550–600 bp were purified and used as templates for PCR 2 using the forward primer VHBACKA6 and reverse primer (F4rev: 5′-ATA GGA TCC GAC CTG GGT CCC CTG GCC CCA-3′) with BamHI recognition site (underlined) added-on at the 3′ end of the gene. Amplicons were pooled and DNAs were purified with phenol chloroform then digested by PstI and BamHI (100 ug) and unidirectionally ligated into PstI- and BamHI-digested pSEX81 (4882 bp) to recover pSEX-VHH. Then, the vector was electroporated into XL1-Blue electrocompetent cells at 200 Ω, 2.5 kV, and 25 μF followed by addition of 0.9 ml SOC media and shaking at 37 °C for 1 h. The library was stored in glycerol at −80 °C until use.

(Color online). Design of stepwise PCR to recover the vhh gene (∼350 bp) from camelids blood sera.
2.3 Library evaluation and sequencing
Twenty microliters of pooled transformants in the library were evaluated by plating on LB selective medium (100 mg Ampicillin/l) for library size. Further, 200 colonies were randomly collected and used for miniprep and the inserts were checked for redundancy in the library by digesting DNAs with HinfI. Then, 100 colonies were sent for capillary DNA sequencing by Macrogen Korea (Gasan-dong, Geumchen-gu, Seoul, Korea). Deduced amino acid sequences were aligned to reference vhh antibody (acc. No. ADE99145) via CLUSTAL W method to detect diversity within VHH region.
3 Results
The stepwise PCR (Fig. 2) was done and recovered amplicons (Fig. 3) were used in the construction of the camelids VHH library. Since this is a naive library (i.e., from non-immunized animals), it was expected to have a high diversity. The size of the library was checked by inoculating 10 ul in replicates of culture plates and counting the number of colonies. The library was estimated to be around 107 cells. Test of redundancy based on digesting DNAs with HinfI enzyme (data not shown) and vhh gene sequencing indicated high diversity among CDR domains of VHH (Fig. 4). The cassette vector pSEX81 was used in the present study as the first step towards the construction of phage display library [21]; the most preferred type of libraries utilized for both disease diagnosis and treatment [22].
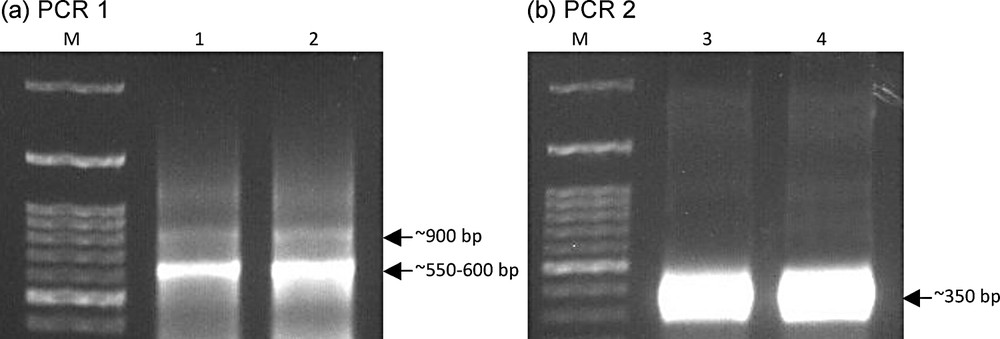
Amplicons of stepwise PCR to recover vhh gene. Amplicons of PCR 1 (a) with ∼550-600 bp sizes (Lanes 1 and 2) of the vhh gene and hinge sequences were cut and gene cleaned. Recovered DNAs were used as templates for PCR 2 (b) to recover ∼350 bp (Lanes 3 and 4) of vhh gene. The design of stepwise PCR and the expected amplicon sizes are illustrated in Fig. 2. M refers to DNA standard (100-bp ladder, Bioron, Germany).
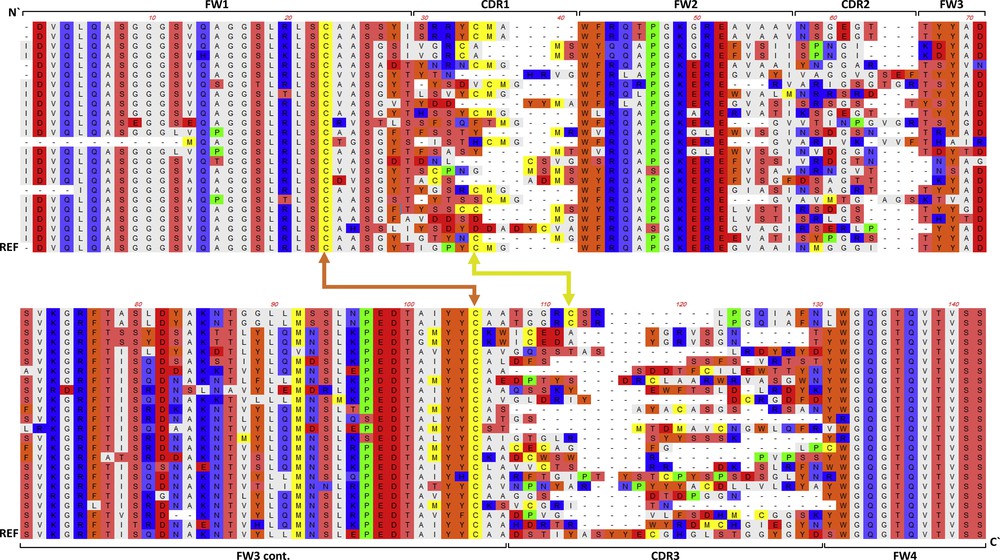
Alignment of the deduced amino acids sequences of the vhh gene isolated from selected colonies of the naïve library. The sequence indicates the four frameworks (FW) and three CDRs. Two disulfide (S) bonds occurring between FW1 and FW3 (orange) and between CDR1 and CDR3 (yellow) are indicated. REF refers to the amino acids sequence of the reference vhh gene.
The vector pSEX81 was designed for the convenient insertion of heavy-chain variable domain coding regions and for production of functional single-chain Fv antibody-pIII fusion proteins on the surface of M13 bacteriophages. The corresponding DNA fragments are usually amplified by PCR. Then, amplicons should be cloned in-frame between a signal peptide sequence of bacterial pectate lyase (pelB) for the secretion of the fusion protein into the periplasmic space, and the pIII gene of M13 bacteriophage. The vector also provides an IPTG inducible strong promoter, the T7 terminator, the ColE1 origin of replication, the intergenic region of phage F1 and an ampicillin resistance marker for selection. In the current work, we have used the recognition sites of the BamHI and PstI restriction endonucleases to allow the unidirectional insertion of vhh gene fragments in-frame with pelB leader and pIII gene (Fig. 5). Now, the vector is ready to be used for the overexpression of any functional recombinant single-chain Fv antibody-pIII fusion proteins in Escherichia coli.
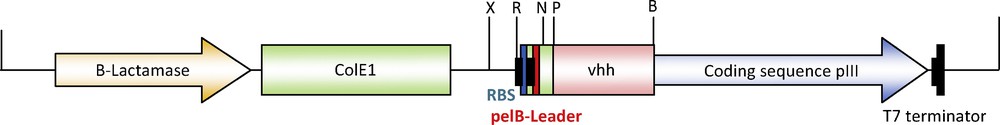
(Color online). Restriction map of pSEX81-VHH indicating the insertion sites of vhh gene (∼350 bp). The gene replaced the ScFv domain in the fusion protein, while retaining the pelB leader for secretion of the fusion protein into the periplasmic space. X: XhoI; R: EcoRI; N: NcoI; P: PstI; B: BamHI.
4 Discussion
Physical connection of light- and heavy-chain variable regions of a conventional antibody results in structural instability, which reduces its solubility and applications. The identification of antibodies formed only by the heavy-chain (HCAbs) in Camelidae and sharks are candidates for the improvement of the antigen – antibody binding [1,2,18] as they are more stable and recognize epitopes inaccessible to conventional Abs [12,23–25].
VHHs recovered so far have mostly been isolated from immunized, rather than non-immunized, camels [1,26]. More recently, naïve VHH libraries from llamas and sharks have been proposed as a better alternative [14–17], where the large diversity of such libraries can allow the identification of binders for any potential antigen. Recent studies indicated the possibility to retrieve useful binders even when using a relatively small naïve library (107) [16], a size similar to that recovered in the present study.
Based on the success to recover the naïve library of 107 size, we propose to recover a similar library with random hypermutations in the CDR domains, a procedure successfully used by many researchers [14,18,27]. These hypermutations can compensate for somatic hypermutation already acquired into libraries recovered from immunized animals [25,28]. Muyldermans et al. [4] showed that a llama naïve library, built from 109 lymphocytes, could recover antibodies with affinity comparable to that of monoclonal antibodies.
In conclusion, the camelids naïve library recovered from the present work can be considered as a valid tool for selecting useful VHHs against different pathogens.
Disclosure of interest
The authors declare that they have no conflicts of interest concerning this article.
Acknowledgements
This project was funded by the Deanship of Scientific Research (DSR), King Abdulaziz University, Jeddah, under Grant No. (1-3-1432/HiCi). The authors, therefore, acknowledge DSR technical and financial support.


