1 Introduction
Stripe (yellow) rust caused by Puccinia striiformis f. sp. tritici (Pst) is the most destructive fungal disease for wheat, especially in cool temperate and humid environment of wheat producing regions like India, China, US, Australia and the middle east [1,2]. Wheat is a widely cultivated cereal crop and acts as staple food to 35% of the world's population, providing 8 to 15% proteins, up to 20% of calories intake and nearly 55% of the carbohydrate [3]. In India, yellow rust (YR) severely affects wheat production in North-Western plain zone as well as Northern hills zone due to the favourable environment for rust pathogens [4,5]. The rust fungus can travel by wind through urediniospores over thousands of kilometres from the initial infection sites [6] and exerts greatest damage by preventing transport of water and nutrients through the host. It causes decrease in seed vigour leading to shrivelled grains, loss in number of grains per spike and of grain weight. Disease loss can range up to 10–70% to complete crop failure [7]. The predominant pathotypes i.e. 46S119 and 78S84 are adversely affecting wheat production in the northern wheat growing region of India [8,9]. A Yellow rust pathotypes profiling in Punjab showed the dominance of race 46S119 in 84% of collected yellow rust samples [10]. Since 2011, the frequency of 46S119 has increased up to 74% whereas that of 78S84 has reduced to 18.5%, due to the favourable cold and humid climate over the years [11]. This shift in virulence and rapid emergence of new races has resulted in breakdown of major resistance genes Yr2, Yr9 and Yr27, rendering widely-planted wheat varieties (PBW 343 and HD 2967) susceptible [11]. YR genes such as Yr5, Yr10, Yr11, Yr12, Yr13, Yr14, Yr15, Yr24, Yr26, YrSp, and Yrsk are still effective and could be used in breeding programs, while Yr2, Yr3, Yr4, Yr6, Yr7, Yr8, Yr9, Yr17, Yr18, Yr19, Yr21, Yr22, Yr23, Yr25, Yr27 and YrA have become ineffective to the currently prevalent or new races [12]. Presently, 76 Yr genes have been catalogued in wheat [13] in which mostly confer race-specific all-stage resistance, and some confer adult-plant or high temperature adult-plant (HTAP) resistance [7,14]. Breeding for resistant cultivars is considered the most effective, economic and environmentally friendly approach to control stripe rust [15,16]. Race-specific resistance with single resistance gene is often short lived. Gene pyramiding strategies deploying race-specific and non-race specific resistance genes is required to achieve durable resistance [17]. However, detailed knowledge of resistance genes present in wheat cultivars is a prerequisite in resistance breeding. Therefore, evaluation and distribution of Yr genes needs to be done in breeding wheat genotypes to identify and select resistant cultivar. Molecular marker assisted detection is the most convenient and reliable method to identify the presence of Yr genes [18,19]. A wide range of markers are reported to be associated with Yr genes in wheat including STS (Yr5- [20]), SSR (Yr10- [21]; Yr15, Yr26, YrH52- [22]), EST (Yr26- [23]), STS/CAPS ([24]; YrMoro- [25]), STS (Yr61- [26]), DArt (Yr51- [27]), RGAP/SSR (Yr59- [28]) and SSR (Yr 50- [29]; Yr64 and Yr65- [30]). Canadian YR resistant wheat varieties were identified with Yr 10, Yr17, Yr18 and Yr 36 linked markers [31]. Ullah et al. [32] screened 99 wheat genotypes using markers linked with Yr5, Yr10, Yr17 and Yr9. Iqbal et al. [33] studied the allelic variation for markers Xgwm 11, CYS5, Xpsp3000, S19M93-140, cSLv34, VENTRIUP/LN2 and Barc181 linked with Yr5, Yr9, Yr10, Yr17, Yr26 in 67 Pakistani wheat varieties. Kumar et al. [34] evaluated 19,460 Indian wheat accessions to identify new sources of resistance to rust using molecular markers. Similar studies were performed using 330 cultivars and 164 breeding genotypes from China [35]. Zheng et al. [36] studied the distribution of 36 Yr genes in 672 wheat accessions in USA and observed significant additive effects in some gene combinations, such as Yr9 + Yr18 and Yr30 + Yr46.
The present study was conducted to explore the rust resistance potential of advanced wheat genotypes and varieties using molecular markers linked with YR resistance genes. The information generated on gene sources of rust resistance will prove vital for developing pre-breeding potential genotypes and high yielding rust resistant varieties. The gene specific rust resistant wheat varieties so developed and deployed would not only reduce cost of cultivation spent on fungicides but would also save environment from pollution.
2 Materials and methods
2.1 Plant materials
A total of 68 wheat genotypes (Table 1) were used for molecular and field evaluation at the Chaudhary Charan Singh Haryana Agricultural University (CCS HAU), Hisar, India.
List of the genotypes used for molecular and field evaluation.
| Sr. No. | Genotype | Sr. No. | Genotype | Sr. No. | Genotype |
| 1 | HI 8759 (d) | 25 | WB 2 | 49 | WH 1184 |
| 2 | PBW 723 | 26 | AKAW 4842 | 50 | HS 580 |
| 3 | HI 8774 (d) | 27 | DBW 179 | 51 | VL 1009 |
| 4 | HPPAU 05 | 28 | DBW 216 | 52 | UP 2954 |
| 5 | HPW 423 | 29 | DBW 217 | 53 | UP 2955 |
| 6 | HPW 433 | 30 | DBW 219 | 54 | VL 3002 |
| 7 | HS 622 | 31 | DDK 1050 (dic.) | 55 | VL 3010 |
| 8 | HS 623 | 32 | DDK 1051 (dic.) | 56 | VL 3011 |
| 9 | HS 626 | 33 | GW 477 | 57 | VL 3012 |
| 10 | HS 628 | 34 | MACS 5044 (dic.) | 58 | WH 1181 |
| 11 | PBW 725 | 35 | MACS 5046 (dic.) | 59 | WH 1216 |
| 12 | PBW 756 | 36 | NW 6094 | 60 | WH 1310 |
| 13 | PBW 757 | 37 | PBW 621 | 61 | HD 3171 |
| 14 | PBW 760 | 38 | RKD 292 (d) | 62 | Lassik |
| 15 | RKD 283 (d) | 39 | VL 4001 | 63 | WH 711 |
| 16 | TL 3006 (T) | 40 | WH 1215 | 64 | PBW 343 |
| 17 | TL 3007 (T) | 41 | DBW 220 | 65 | WH 542 |
| 18 | TL 3008 (T) | 42 | HPBW 02 | 66 | WH 1105 |
| 19 | TL 3009 (T) | 43 | HPPAU 08 | 67 | HD 2967 |
| 20 | UAS 459 (d) | 44 | HPPAU 10 | 68 | Kharchia |
| 23 | HI 1605 | 47 | NW 6046 | ||
| 24 | K 1317 | 48 | PDW 344 (d) |
2.2 Molecular markers
Two or three closely linked markers of each YR gene were chosen to identify its presence or absence in the wheat genotypes, except a few genes for which only one closely linked marker was reported. A total of 70 markers (SSR, CAPS, RGAP, STS, EST-SSR) were used for profiling the wheat genotypes. Sequences of available markers (Table 2) along with their previously determined chromosomal locations were obtained from Graingenes database (http://wheat.pw.usda.gov/). Primers were synthesized from Eurofins Genomics India Pvt. Ltd, Bangalore and Imperial life sciences Pvt. Ltd. India.
Primers’ name, sequences, distance, location, product size, annealing temperature, and references for stripe rust resistance genes.
| Gene | Linkage | Chromosome position | Source genotypes | Type of resistance conferred | Marker name | Type of Marker | Primer sequence | Annealing Temperature | Distance (cm) | Product size | References |
| Yr2 | 7B | RS | Wmc364 | SSR | ATCACAATGCTGGCCCTAAAAC CAGTGCCAAAATGTCGAAAGTC |
56 | 5.6 | 200 (+), 190 (–) | Feng et al. [113] | ||
| Yr5 | Yr7 (allelic or linked) | 2B | Triticum aestivum subsp. spelta ‘Album’ | RS, AS |
Wmc175
|
SSR | GATAAAATCATTATTGGGTGTCCTTT TTCAAATAATCTTTCATCAGTCAAATG |
55 | 1.1 | Not amplified | Sun et al. [46] |
| Xgwm501 | SSR | ACTTACATGAATTATCTTTCTTGGTCC CGTATTCAAATAATCTTTCATCAGTCA |
55 | 10.5–13.3 | 100, 150, 176 (+) | Sun et al. [46] | |||||
| STS7/8 | STS | GTACAATTCACCTAGAGT GCAAGTTTTCTCCCTATT |
45 | 0.3 | 500 (+) | Murphy et al. [48] | |||||
| S19M93 | STS | TAATTGGGACCGAGAGACG TTCTTGCAGCTCCAAAACCT |
58 | 0 | 100 (+) | Smith et al. [47] | |||||
| Barc349 | SSR | CGAATAGCCGCTGCACAAG TATGCATGCCTTTCTTTACAAT |
58 | 0.4 | 100, 105 (+), 120, 140, 220 | Murphy et al. [48] | |||||
| Barc167 | SSR | AAAGGCCCATCAACATGCAAGTACC CGCAGTATTCTTAGTCCCTCAT |
55 | 2.6 | Not amplified | Smith et al. [47] | |||||
| Yr7 | Sr9 g | 2BL | T. turgidum (lumillo durum) | RS, AS | Xgwm526 | SSR | CAATAGTTCTGTGAGAGCTGCG CCAACCCAAATACACATTCTCA | 55 | 5.6 | 75, 78, 95, 140, 145 (+) | Yao et al. [51] |
| CFD77 | SSR | CTGCTTCAGGGATTGGAGAG GTTTCCTGGGCTAAACCACA |
58 | 220 | USDA | ||||||
| Yr9 | Sr31/Lr26/Sr50 | 1RS/1BL | Secalis cereal (Imperial rye) | RS, AS | Xgwm582 | SSR | AAGCACTACGAAAATATGAC TCTTAAGGGGTGTTATCATA |
45 | 3.7 | 150 (+), null | Cabuk et al. [54] |
| STS (Sr31) | STS | CTCTGTGGATAGTTACTTGATCGA CCTAGAACATGCATGGCTGTTACA |
58 | 0 | Not amplified | USDA | |||||
| IB-267 | SSR | GCAAGTAAGCAGCTTGATTTAGC AATGGATGTCCCGGTGAGTGG | 56 | 0 | 267 (+), null | Mago et al. [55] | |||||
| Xwgp8 | RGAP | CTCTGTATACGAGTTGTC GAGGAAGGACAGGTTGCC |
60 | Not amplified | USDA | ||||||
| Yr10 | 1BS | ‘Moro’ | RS, AS |
Xpsp3000
|
SSR |
GCAGACCTGTGTCATTGGTC GATATAGTGGCAGCAGGATACG |
52 | 1.5 |
240 (–), 220 (+), 260 (+), 285 (+) |
Bariana et al. [60] | |
| Yr15 |
YrCH52
(close) |
1BS | T. turgidum var. dicoccoides G-25 | RS, AS | Xgwm413 | SSR | TTTTTGGCTTATTAGACTGACTT TTGCCATAAAATACAAAATCC |
50 | 4.3 | 90, 95 (+), 100 (–) | Murphy et al. [48] |
| Xgwm11 | SSR | AAAAGGAACCTCAAGTGACA GAAAATGAGGGAGTGAGATG |
52 | Not amplified | Cheng et al. [30] |
||||||
|
Xgwm273
|
SSR | ATTGGACGGACAGATGCTTT AGCAGTGAGGAAGGGGAT C |
58 | 0.4 | 156 (+), 165 170, 180, 190, 200 |
Revathi et al. (2010) [114] | |||||
| Xbarc8 | SSR | GCGGGAATCATGCATAGGAAAACAGAA GCGGGGGCGAAACATACACATAAAAACA |
56 | 4.2 | 260 (+), 280 | Murphy et al. [48] |
|||||
|
Xgwm18
|
SSR | TGGCGCCATGATTGCATTATCTC GGTTGCTGAAGAACCTTATTTAGG | 52 | Not amplified | Cabuk et al. [54] | ||||||
| Xgwm33 | SSR | GGAGTCACACTTGTTTGTGCA CACTGCACACCTAACTACCTGC |
55 | 4.5 | Not amplified | Somers et al. [128] | |||||
| Yr16 | 2AS, 2DS | Capelle-Desprez | NRS, AP |
Xgwm102
(QTL) |
SSR |
TCTCCCATCCAACGCCTC TGTTGGTGGCTTGACTATTG | 57 | 150 (–), 155 (+), 200 (+) | Agenbag et al. [63] | ||
| Xgwm 249 | SSR | CAAATGGATCGAGAAAGGGA CTGCCATTTTTCTGGATCTACC |
58 | 120–160 | Agenbag et al. [63] | ||||||
| Yr17 | Lr37/Sr38 | 2AS | T. ventricosa | RS, AS |
VENTRIUP-LN2 |
AGGGGCTACTGACCAAGGCT TGCAGCTACAGCAGTATGTACACAAAA |
58 |
T. ventricosum
chromosome specific |
259 (+), null |
Halguera et al. (2003) [24] |
|
| URIC/LN2 | CAPS | GGTCGCCCTGGCTTGCACCT TGCAGCTACAGCAGTATGTACACA AAA |
58 | 285 (+), 166 (–), 109 (–) | Halguera et al. (2003) [24] | ||||||
| Yr18 | Lr34 | 7D | Frontana | NRS, AP | csLV34 | GTTGGTTAAGACTGGTGATGG TGCTTGCTATTGCTGAATAGT |
58 | 0.4 | 150 (+), 229 (–) | Lagudah et al. [65] | |
| Cssfr1 | Gene based | TTGATGAAACCAGTTTTTTTTCTA GCCATTTAACATAATCATGATGGA |
56 | 0 | Not amplified | Lagudah et al. [129] | |||||
| Cssfr2 | Gene based | TTGATGAAACCAGTTTTTTTTCTA TATGCCATTTAACATAATCATGAA |
55 | 0 | 517 (+), null | Lagudah et al. [129] | |||||
| Cssfr5 | Gene based | TTGATGAAACCAGTTTTTTTTCTA TATGCCATTTAACATAATCATGAA |
57 | 0 | Not amplified | Lagudah et al. [129] | |||||
| Xgwm295 | SSR | GTGAAGCAGACCCACAACAC GACGGCTGCGACGTAGAG |
55 | Variable bands | Bariana et al. [117] | ||||||
| CHR5 | RGAP | GCATTGGAACAAGGTGAA GGIGGIGTIGGIAAIACIAC |
0 | Not amplified | Wen et al. [69] | ||||||
| Yr24 | Yr24, YrCH42 | 1BS | T. turgidum | RS, AS | Barc181 | SSR | CGCTGGAGGGGGTAAGTCATCAC CGCAAATCAAGAACACGGGAGAAAGAA |
58 | 6.7 | 180 (+), 220 (–) | Wang et al. [68]; Zhang et al. [23] |
| Yr25 | 1D | RS, AS | Xgwm6 | SSR | CGTATCACCTCCTAGCTAAACTAG AGCCTTATCATGACCCTACCTT |
52 | 150 | [131] | |||
| Yr26 | Yr24, YrCH42 | 1BS | Haynaldia villosa | RS, AS | Barc187 | SSR | GTGGTATTTCAGGTGGAGTTGTTTTA CGGAGGAGCAGTAAGGAAGG |
57 | 2.3 | 200 (+), 220 (–), 225 | Wen et al. [69] |
| CON-6 (EST) | ESTSSR | GCCGATGGGGAACTGAAT GTTGAACCGCTTGAACACC |
52 | 0 | Not amplified | Zhang et al. [23] | |||||
| CYS-5 | RGAP | GCATTGGAACAAGGTGAA GGIGGIGTIGGIAAIACIAC |
58 | 0 | Not amplified | Wen et al. [69] | |||||
| Xgwm498 | SSR | GGTGGTATGGACTATGGACACT TTTGCATGGAGGCACATACT |
60 | 1.6 | 160 (+), null | Li et al. [67] | |||||
| Yr27 | Lr13 | 2BS | Selkirk | RS, AS | Xgwm630 | SSR | GTGCCTGTGCCATCGTC CGAAAGTAACAGCGCAGTGA |
55 | 10 | 124 (+) | Seyfarth et al. (2000) [119] |
| Yr29 | Lr46, YrChk | 1BL | Lalbahadur | NRS, AP | Xgwm259 | SSR | AGGGAAAAGACATCTTTTTTTTC CGACCGACTTCGGGTTC |
56 | (Yrchk: 9.1) | Not amplified | Lilemo et al. (2008) [120] |
| Wmc44 | SSR | GGTCTTCTGGGCTTTGATCCTG TGTTGCTAGGGACCCGTAGTGG |
58 | 3.6 (Yrchk: 8.3) |
205, 210, 220, 240, 260 (+), 280, 300, 320 | Rosewarne et al. [75]; Liu et al. (2007) [121] |
|||||
| Yr30 | Sr2, Lr27 | 3BS | Opata 85 | NRS, AP | Xgwm533 | SSR | AAGGCGAATCAAACGGAATA GTTGCTTTAGGGGAAAAGCC |
57 | 100, 120 (+), 130, 140, 150 | Spielmeyer et al. (2003) [122] | |
|
csSr2
|
CAPS | CAAGGGTTGCTAGGATTGGAAAACA AGATAACTCTTATGATCTTACATTTTTCTGA |
56 | 0 | Not amplified | Mago et al. (2011) [123] | |||||
| Yr32 | 2AL | Carstens V | RS, AS | Wmc198 | SSR | CACGCTGCCATCACTTTTAC TTGAAGTGGTCATTGTTGCT |
58 | 2 | 180, 200 | Eriksen et al. [80] | |
| Yr35 | Lr53 | 6BS | T. dicoccoides | RS, AS | CFD1 | SSR | CCTCCATGTAGGCGGAAATA TGTGTCCCATTCACTAACCG | 57 | 4.1 | Not amplified | Dadkhodaie et al. [132] |
| Yr36 | Gpc-B1 | 6BS | T. dicoccoides | NRS, HTAP | Barc101 | SSR | GCTCCTCTCACGATCACGCAAAG GCGAGTCGATCACACTATGAGCCAATG |
58 | 2 | 116, 124 (+), 138, 160, 165 | Uauy et al. [82] |
| ASA (Gpc-B1) | Gene based | CTACCATCGAAAGTTGATAGGGA TTCACAAACTAAGGGGAGGGA |
57 | 0.3 | 1.6Kb (+), null | Uauy et al. [82] | |||||
| WKS1 | Gene based | ATCCATTGCCAAGTCAACCAC TCACTTCCATGAAGGAGGTC |
55 | 0 | 128 (+), null | Fen et al. (2009) [83] Fu et al. (2009) [[130]] |
|||||
| Yr39 | 7BL | Alpowa | NRS, HTAP |
Xwgp36:
Pto kin1/RLK For |
RGAP |
GCATTGGAACAAGGTGAA GAYGTNAARCCIGARAA |
55 | 0.8 | Not amplified | Lin and Chen [115] | |
| Xwgp45:Pto kin1/XLRR For | RGAP | GCATTGGAACAAGGTGAA CCGTTGGACAGGAAGGAG |
52 | 6.6 | Not amplified | ||||||
| Yr40 | Lr57 | 5DS | Aegilops geniculata | RS, AS | Xfbb276 | SSR | AACAGCTATGACCATG GTAAAACGACGGCCAGT |
48 | 1 kb, 2 kb, null | Kuraparthy et al. [84] | |
| Yr45 | 3DL | RS, AS |
Xwgp115
|
RGAP | AGTGTCTTGTAGGGTATC TCAGGCCGTGAAAAATAT |
46 | 4.8 | Not amplified | Li et al. (2011) [124] | ||
| Xwgp118 | RGAP | AAGTGGAACAAGGTTACG ACACTGGTCCATGAGGTT |
48 | 5.8 | Not amplified | Li et al. (2011) [124] | |||||
| Yr46 | Lr67/Sr55/Pm46 | 4DL | Thatcher | RS, AS | CFD71 | SSR | CAATAAGTAGGCCGGGACAA TGTGCCAGTTGAGTTTGCTC |
52 | 148 (+), 150, 152 | Hiebert et al. (2010) [125] | |
| CFD23 | SSR | TAGCAGTAGCAGCAGCAGGA GCAAGGAAGAGTGTTCAGCC |
55 | 211 (+), 214 (+) | Hiebert et al. (2010) [125] | ||||||
| Yr47 | Lr52 | 5BS | T. aestivum | RS, AS |
Cfb309
|
SSR (BAC Contig) |
TAGGGCATATTTCCAACACT TAAGTCCGCGTATTAGCATT |
58 | 8–12 | 600 (+), 350 (–) | Bansal et al. [86] |
| Yr51 | 4AL | RS, AS | Sun104 | STS | TGCTATGTGCGTGATGATGA TTACATGCTCCAGCGACTTG |
56 | 2.5 | 225 (+), null (–) | Randhawa et al. [27] | ||
|
owm45F3R3
|
SSR | GGCTCGTCTACACCAACGAC TTGGGGTCTTTAGGCATGAG |
56 | 1.2 | 1 kb (non conclusive) | Randhawa et al. [27] | |||||
| Yr59 | 7BL | Alpowa | NRS, HTAP | Barc32 | SSR | GCGTGAATCCGGAAACCCAATCTGTG TGGAGAACCTTCGCATTGTGTCATTA |
62 | 2.1 | 165 (+), 175, 190, 250 | Zhou et al. [26] | |
| Wmc557 | SSR | GGTGCTTGTTCATACGGGCT AGGTCCTCGATCCGCTCA |
56 | < 2.1 | 315 (+), 500 | Zhou et al. [26] | |||||
| Yr60 | 4AL | T. aestivum | RS, AS | Wmc776 | SSR | CCATGACGTGACAACGCA ATTGCAGGCGCGTTGGTA |
56 | < 0.6 | 150, 160, 170 (+) | Herrera-Foessel et al. [85] | |
| Wmc313 | SSR | GCAGTCTAATTATCTGCTGGCG GGGTCCTTGTCTACTCATGTCT |
60.3 | 0.6 | 180, 200 (+) | Herrera-Foessel et al. [85] | |||||
| Wmc219 | SSR | TGCTAGTTTGTCATCCGGGCGA CAATCCCGTTCTACAAGTTCCA |
57 | 0.6 | 200 (+), 220 | Herrera-Foessel et al. [85] | |||||
| YrCH52 | Yr15 | 1BS | T. turgidum | RS, AS | Xgwm273 | SSR | ATTGGACGGACAGATGCTTT AGCAGTGAGGAAGGGGATC | 48 | 2.7 | 170, 180, 190, 200 | Cabuk et al, (2011) [54] |
| YrSpP | 2B | Spaldings Profilic | RS, AS | Wmc441 | SSR | TCCAGTAGAGCACCTTTCATT ATCACGAAGATAAACAAACGG | 56 | 12.1 | Not amplified | Guan et al. (2005) [116] | |
| Yr48 | 5AL | Aegilops tauschii | NRS, AP | cfa2149 | SSR | CTT GGA GCT CGG GTA GTA GC AAG GCA GCT CAA TCG GAG TA |
52 | 0.06 | 225 (+), 250 | Lowe et al. (2011) [126] | |
| SNF-A2 | SSR | TCCGTCTCCATCATTCAACA GTGTTGCGCAAGTTTGTGAC |
58 | 0.18 | 150 (+), 180, 200 (–) |
Lowe et al. (2011) [126] | |||||
| BE495011 | SSR | TGATTACTGTAGCTACCTCCTCCT GGTGCAAGATGTGCCTGTAA |
56 | 0.09 | 220 (+), null | Lowe et al. (2011) [126] | |||||
| YrCN19 | 2BS | AIM6 | RS, AS | Xgwm410 | SSR | GCTTGAGACCGGCACAGT CGAGACCTTGAGGGTCTAGA |
57 | 0.3 | Not amplified | Luo et al. (2008) [117] | |
| YrZH84 | 7B | RS, AS | Barc32 | SSR | GCGTGAATCCGGAAACCCAATCTGTG TGGAGAACCTTCGCATTGTGTCATTA |
56 | 4.8 | 165 (+), 170, 190, 250 |
Zhou et al. ([26]; [125]) | ||
| YrMor | 4B | Moro | RS, AS | S26M47 | SSR | TTTACAGGTTGGAATCTA GAATATACCTTTTCTTCAA |
55 | 0 | 250 (+), null | Smith et al. [25] | |
| YrHua | RS, AS | PM14 | STS | GTACATGCAGACAGAAAGAGAGAA TGATGAGTCCTGAGTAACTC |
58 | 5.4 | Not amplified | Cao et al. (2008) [127] | |||
| YrExp1 | RS, AS | Wmc631 | SSR | TTGCTCGCCCACCTTCTACC GGAAACCATGCGCTTCACAC |
60 | 3.4 | Not amplified | Lin and Chen (2008) [118] |
2.3 DNA extraction, PCR amplification and electrophoresis
Genomic DNA was isolated from 100 mg fresh leaf tissue collected from each line using the CTAB extraction method of Murray and Thompson [37] as modified by Saghai-Maroof et al. [38] and Xu et al. [39]. The DNA stock solution was adjusted to a concentration of 100–150 ng/μl with nuclease free sterile water as the working concentration for the polymerase chain reaction (PCR) and stored at -20° C. PCR reactions were performed using a Benchtop thermocycler in 25 μl of a PCR mixture containing 100–150 ng of genomic DNA, 2 units of Taq DNA polymerase (Promega), 1X PCR buffer (10 mM Tris HCL), 2.5 mM of MgCl2, 100 μM of each dNTP, and 10 μM of each primer. The Touchdown PCR protocol was used to increase the specificity and sensitivity in PCR amplifications. The annealing temperature for Touchdown PCR was optimized using the melting temperature recommended by the primer synthesis user's guide. The initial program provided for a decrease of 0.5 °C for 40 s per 5 cycles from 65 to 60 °C; the second one started at 60 °C with a decrease of 0.5 °C for 40 seconds per 30 cycles. PCR products were separated on 1.5–2.5% agarose gels (depending on gene product size) or 6% denaturing polyacrylamide gel and visualized under UV light using the digital gel imaging system (MultiDoc-It). For the primer pair URIC/LN2, the PCR products were digested with Dpn II (New England Biolabs, USA) according to Chen et al. [20] before electrophoresis. The restriction digestion mixture contained 10 μl of PCR product, 1U of restriction enzyme Dpn II (New England Biolabs) and 2 μl of 10 X buffer for Dpn II (New England Biolabs). Samples were incubated at 37 °C for 4 h and the digested products were separated in a 6% polyacrylamide gel.
2.4 Field evaluation under artificial inoculation of Puccinia striiformis f. sp. tritici for stripe rust
All 68 wheat genotypes were evaluated under epiphytotic condition by artificial inoculation of mixture of pathotypes including 46S119, 110S119, 110S84 & 78S84 (106/ml) at seedling/tillering stage of Pst through spray for stripe rust reaction in the research field of CCS HAU, Hisar (latitude 29°10 ’N, longitude 75°46 ’E, altitude 215.2 m) in India during the 2016–17 cropping season. Sowing was done in mid November. Approximately, 10 g seeds of each of the 68 wheat genotypes were planted in 2 m furrow length with a distance of 25 cm between each furrow in the field. The infectors (mixture of susceptible cv. i.e. Kharchia, Agra Local, WH143, Lal Bahadur & Bajara Yellow) were sown after every 20 furrows and around the plots as natural spreaders of stripe rust inoculum. Infection types (ITs) of seedlings were recorded in late January and early February when the plants were at tillering/stem elongation stage and disease severity (DS) data was also recorded. In late March, terminal disease severity was also recorded using modified Cobb's scale [40]. The immune plant with no visible symptoms was scored as IT-0, the highly resistant plant with little necrotic flecks and no sporulation was scored as IT-1, the moderately resistant plant with a few necrotic flecks and trace sporulation was scored as IT-2, the moderately susceptible plant with necrotic blotches and moderate sporulation was scored as IT-3, the highly susceptible plant with chlorotic stripes and abundant sporulation was scored as IT-4. Disease severities were assessed based on the percentage of leaf area affected (0, 1, 5, 10, 20, 30, 40, 60, 80, and 100%) [41]. Disease severity were recorded as R (Resistant) when small necrotic areas and no uredia were present, MR (moderately resistant) when small uredia with slight sporulation, chlorosis or necrosis, MS (moderately susceptible) when medium-size uredia with moderate sporulation, and some chlorosis may still be present, and S (susceptible) when large uredia with abundant sporulation, and often coalesced to form lesions without evidence of stripes on visible chlorosis or necrosis [42].
3 Results and discussion
3.1 Adult-plant stage rust evaluation in field under artificial conditions
At the adult-plant stage, disease data on infection types and severity was recorded on 68 wheat genotypes as depicted in Tables 3 and 4. Fifty-three genotypes were found resistant (ITs 0), accounting for 77.94% of total genotypes, 3 genotypes (4.4%) (RKD 292, VL 3002, and DBW 179) showed trace resistance (ITs 1), 7 genotypes (10.3%) (HD 3209, AKAW 4842, DDK 1050, GW 477, MACS 5044, MACS 5046 and NW 6094) were moderately susceptible (ITs 3) and 5 genotypes (7.35%) (HD 3171, DDK 1051, Infector, WH 711 and PBW 343) were susceptible (ITs 4). According to disease severity, 53 lines exhibited immune response, 7 genotypes (10.3%) (HD 3209, AKAW 4842, DDK 1050, GW 477, MACS 5044, MACS 5046 and NW 6094) showed 10 percentage severity of moderately susceptible type (10MS), one genotype (1.5%) HD 3171 contracted 5 percentage severity of susceptible type (5S), 4 genotypes (5.9%) (DDK 1051, WH 711, PBW 343 and Infector) obtained 60 percentage severity of susceptible type (60S). Infector rows expressed 60 percentage severity of susceptible type (60S). Coefficient of infection ranged from 0 to 60 among all wheat genotypes in which 0 CI is associated with the immune and resistant genotypes and 60 CI is associated with susceptible genotypes. Overall 60 genotypes expressed yellow rust resistance under field conditions against predominant yellow rust pathotypes i.e. 46S119, 110S119, 110S84 &78S84.
Field response, severity, and coefficient of infection of wheat genotypes.
| Genotypes | Field Severity response & symbol | No. of Lines | Average Coefficient of Infection (ACI) |
| RKD 292, VL3002, DBW179 | Ts (R) | 3 | 0.2 |
| HD 3209, AKAW 4842, DDK 1050, GW 477, MACS 5044, MACS 5046, NW 6094 | 10MS | 7 | 8.0 |
| HD 3171 | 5S | 1 | 5.0 |
| DDK1051, Infector, WH 711, PBW 343 | 60S | 4 | 60.0 |
| Remaining genotypes | Immune | 53 | 0 |
Data on infection types (ITs) in the selected wheat genotypes.
| Wheat genotypes | Infection response (field) | Total No. of genotypes | ITs |
| RKD 292, VL3002, DBW179 | Ts | 3 | 1 |
| HD3209, AKAW4842, DDK 1050, GW477, MACS 5044, MACS 5046, NW6094 | MS | 7 | 3 |
| HD3171, DDK1051, Infector, WH711, PBW343 | S | 5 | 4 |
| All remaining genotypes | R | 53 | 0 |
3.2 Detecting stripe (yellow) rust resistance genes using molecular markers
Marker assisted detection of 35 Yr genes in wheat genotypes was carried out using 70 Yr gene linked markers. The results indicated the effectiveness of these markers for specific Yr gene which also identified the resistant lines containing multiple Yr genes. Details of Yr resistance genes in wheat genotypes are presented in Table 5. Molecular identification of various genes is discussed below.
Details of Yr resistance genes in wheat genotypes.
| S. no. | Genotypes | RS, AS genes | APR genes | Total number of RS, AS genes | Total number of APR genes | Total number of genes | Resistant Types |
| 1 | HI 8759 (d) | Yr2, Yr7, Yr10, Yr15, Yr24, YrcH52, Yr47, YrZH84 | Yr16, Yr18, Yr29, Yr30, Yr48, Yr59 | 8 | 6 | 14 | R |
| 2 | PBW 723 | Yr2, Yr9, Yr24, YrZH84 | Yr16, Yr30, Yr59 | 4 | 3 | 7 | R |
| 3 | HI 8774 (d) | Yr2, Yr7, Yr10, Yr24, Yr40, Yr60 | Yr18, Yr30, Yr48 | 6 | 3 | 9 | R |
| 4 | HPPAU 05 | Yr2, Yr7, Yr9, Yr10, Yr15, Yr24, Yr26, Yr64, YrMor | Yr18, Yr30, Yr36 | 9 | 3 | 12 | R |
| 5 | HPW 423 | Yr2, Yr10, Yr24, Yr26, Yr64 | Yr18, Yr30 | 5 | 2 | 7 | R |
| 6 | HPW 433 | Yr2, Yr7, Yr10, Yr26, Yr47, YrMor | Yr18, Yr48 | 6 | 2 | 8 | R |
| 7 | HS 622 | Yr2, Yr7, Yr24, Yr26 | Yr30 | 4 | 1 | 5 | R |
| 8 | HS 623 | Yr2, Yr7, Yr26, YrMor | Yr18, Yr29, Yr30 | 4 | 3 | 7 | R |
| 9 | HS 626 | Yr5, Yr7, Yr10, Yr24, Yr26, Yr40, Yr47, Yr64 | Yr16, Yr29, Yr30 | 8 | 3 | 11 | R |
| 10 | HS 628 | Yr2, Yr7, Yr15, YrcH52, Yr64 | Yr30 | 5 | 1 | 6 | R |
| 11 | PBW 725 | Yr2, Yr15, Yr24, Yr26, Yr64 | Yr29, Yr30 | 5 | 2 | 7 | R |
| 12 | PBW 756 | Yr2, Yr10, Yr64 | Yr30 | 3 | 1 | 4 | R |
| 13 | PBW 757 | Yr2 | Yr59 | 1 | 1 | 2 | R |
| 14 | PBW 760 | Yr2, Yr7, Yr24, YrMor | Yr59 | 4 | 1 | 5 | R |
| 15 | RKD 283 (d) | Yr7, Yr10, Yr24, Yr64 | Yr59 | 4 | 1 | 5 | R |
| 16 | TL 3006 (T) | Yr24, Yr64 | Yr18, Yr30 | 2 | 2 | 4 | R |
| 17 | TL 3007 (T) | Yr5, Yr10, Yr15, Yr26, Yr40, Yr47 | Yr18, Yr29, Yr30, Yr59 | 6 | 4 | 10 | R |
| 18 | TL 3008 (T) | Yr10, Yr26, Yr64, YrMor | Yr29 | 4 | 1 | 5 | R |
| 19 | TL 3009 (T) | Yr10, Yr24, Yr26, Yr47, YrZH84 | Yr29, Yr48, Yr59 | 5 | 3 | 8 | R |
| 20 | UAS 459 (d) | Yr10, Yr26, Yr64 | Yr29, Yr48 | 3 | 2 | 5 | R |
| 21 | INFECTOR | Yr46 | 0 | 1 | 1 | 30S | |
| 22 | HD 3209 | Yr5, Yr15, Yr26, Yr47 | Yr29, Yr30 | 4 | 2 | 6 | ‘10MS |
| 23 | HI 1605 | Yr10, Yr24, Yr26, Yr40, Yr64 | 5 | 0 | 5 | R | |
| 24 | K 1317 | Yr5, Yr10, Yr24, Yr26, Yr40, Yr64 | Yr30, Yr46 | 6 | 2 | 8 | R |
| 25 | WB 2 | Yr2, Yr5, Yr7, Yr10, Yr26, YrcH52, Yr64 | Yr29, Yr30 | 7 | 2 | 9 | R |
| 26 | AKAW 4842 | Yr7, Yr15, Yr24, Yr26, YrcH52, Yr32, Yr40, Yr64 | Yr36 | 8 | 1 | 9 | 10MS |
| 27 | DBW 179 | Yr2, Yr7, Yr15, Yr24, YrcH52, Yr40, Yr47, Yr64 | Yr16, Yr29 | 8 | 2 | 10 | TS |
| 28 | DBW 216 | Yr2, Yr7, Yr10, Yr15, Yr26, YrCH52, Yr32, Yr40, Yr64, YrMor | Yr29 | 10 | 1 | 11 | R |
| 29 | DBW 217 | Yr2, Yr7, Yr15, Yr24, Yr26, YrCH52, Yr40, Yr47, Yr64 | Yr16, Yr30 | 9 | 2 | 11 | R |
| 30 | DBW 219 | Yr2, Yr10, Yr15, Yr24, Yr26, Yr32 | Yr16, Yr29, Yr46 | 6 | 3 | 9 | R |
| 31 | DDK 1050 (dic.) | Yr2, Yr7, Yr15, Yr24, Yr26, Yr40, YrMor, YrZH84 | Yr29, Yr30, Yr59 | 8 | 3 | 11 | 10MS |
| 32 | DDK 1051 (dic.) | Yr2, Yr5, Yr7, Yr24, Yr26, YrcH52, Yr47, YrZH84 | Yr16, Yr29, Yr30 | 8 | 3 | 11 | 10MS |
| 33 | GW 477 | Yr2, Yr10, Yr15, Yr24, YrcH52 | Yr30, Yr63 | 5 | 2 | 7 | 10MS |
| 34 | MACS 5044 (dic.) | Yr2, Yr15, Yr24, YrCH52, Yr47, Yr64 | Yr16, Yr18, Yr29, Yr46, | 6 | 4 | 10 | 10MS |
| 35 | MACS 5046 (dic.) | Yr2, Yr10,Yr15, Yr24, YrcH52, Yr47, Yr64 | Yr18, Yr29 | 7 | 2 | 9 | 10MS |
| 36 | NW 6094 | Yr2, Yr10, Yr24, YrcH52, Yr47, Yr64 | Yr18, Yr48 | 6 | 2 | 8 | R |
| 37 | PBW 621 | Yr2, Yr7, Yr10, Yr15, Yr24, Yr26, YrcH52,Yr64 | Yr16, Yr30, Yr48 | 8 | 3 | 11 | TS |
| 38 | RKD 292 (d) | Yr2, Yr7, Yr10,Yr15, Yr24, Yr26, YrcH52, Yr47, YrZH84, Yr64,YrMor | 11 | 0 | 11 | R | |
| 39 | VL 4001 | Yr2, Yr10,Yr15, Yr24, YrcH52, Yr40, Yr47, Yr63, YrMor | Yr30 | 9 | 1 | 10 | R |
| 40 | WH 1215 | Yr2, Yr15, Yr24, Yr26, YrcH52, Yr47, Yr64 | Yr46 | 7 | 1 | 8 | R |
| 41 | DBW 220 | Yr2, Yr10, Yr15, Yr26, YrCH52, Yr40, Yr64 | Yr16, Yr18, Yr30, Yr46 | 7 | 4 | 11 | R |
| 42 | HPBW 02 | Yr2, Yr7, Yr10, Yr15, Yr17, Yr64 | Yr18, Yr46 | 6 | 2 | 8 | R |
| 43 | HPPAU 08 | Yr2, Yr10, Yr24, Yr26, YrcH52, Yr64, YrMor | Yr16, Yr18, Yr29, Yr30 | 7 | 4 | 11 | R |
| 44 | HPPAU 10 | Yr2, Yr15, Yr24, Yr26, Yr64 | Yr16, Yr18, Yr29, Yr30 | 5 | 4 | 9 | R |
| 45 | HPW 424 | Yr2, Yr15, Yr24, Yr26, Yr63, YrMor | Yr18, Yr29, Yr30 | 6 | 3 | 9 | R |
| 46 | HS 627 | Yr2, Yr7, Yr10, Yr15, Yr26, Yr64 | Yr29, Yr30 | 6 | 2 | 8 | R |
| 47 | NW 6046 | Yr5, Yr7, Yr10, Yr24, Yr26 | Yr29, Yr30 | 5 | 2 | 7 | R |
| 48 | PDW 344 (d) | Yr7, Yr10, Yr24, Yr26, YrcH52 | Yr29, Yr30, Yr48 | 5 | 3 | 8 | R |
| 49 | WH 1184 | Yr2, Yr5, Yr7, Yr10, Yr26, YrcH52, Yr40, Yr64, YrMor | Yr29, Yr30, Yr48 | 9 | 3 | 12 | R |
| 50 | HS 580 | Yr2, Yr5, Yr7, Yr10, Yr26, YrcH52, Yr40, Yr64 | Yr18, Yr29, Yr30, Yr46, | 8 | 4 | 12 | R |
| 51 | VL 1009 | Yr2, Yr7, Yr10, Yr26, YrcH52, Yr32, Yr40, Yr64, YrZH84 | Yr16, Yr18, Yr29, Yr30, Yr46, Yr59 | 9 | 6 | 15 | R |
| 52 | UP 2954 | Yr2, Yr7, Yr10, Yr24, Yr26, YrcH52, Yr64, YrZH84 | Yr16, Yr29, Yr46, Yr59 | 8 | 4 | 12 | R |
| 53 | UP 2955 | Yr2, Yr7, Yr24, YrcH52, Yr47, Yr48, Yr64 | Yr16, Yr46 | 7 | 2 | 9 | R |
| 54 | VL 3002 | Yr5, Yr7, Yr24, Yr26, YrcH52, Yr40, Yr47, Yr48, Yr60, Yr64 | Yr16, Yr18, Yr29, Yr30, Yr46 | 10 | 5 | 15 | Ts |
| 55 | VL 3010 | Yr2, Yr5, Yr7, Yr10, Yr15, Yr26, YrCH52, Yr40, Yr47 | Yr16, Yr18, Yr29, Yr30, Yr46 | 9 | 5 | 14 | R |
| 56 | VL 3011 | Yr5, Yr7, Yr10, Yr26, YrCH52, Yr40 | Yr18, Yr29, Yr30, Yr46 | 6 | 4 | 10 | R |
| 57 | VL 3012 | Yr2, Yr5, Yr10 | Yr29, Yr30, Yr46 | 3 | 3 | 6 | R |
| 58 | WH 1181 | Yr2, Yr5, Yr7, Yr10, Yr64 | Yr16, Yr29, Yr30 | 5 | 3 | 8 | R |
| 59 | WH 1216 | Yr7, Yr10, YrCH52, Yr64, YrMor | Yr16, Yr30 | 5 | 2 | 7 | R |
| 60 | WH 1310 | Yr2, Yr7, Yr10, Yr40 | Yr16, Yr30, Yr48 | 4 | 3 | 7 | R |
| 61 | HD 3171 | Yr2, Yr7, Yr10, Yr40, YrMor | Yr16, Yr30 | 5 | 2 | 7 | R |
| 62 | Lassik | Yr17, Yr26 | Yr18, Yr29, Yr30, Yr36, Yr48 | 2 | 5 | 7 | R |
| 63 | WH711 | Yr16, Yr30 | 0 | 2 | 2 | S | |
| 64 | PBW343 | Yr2, Yr26 | Yr16, Yr30 | 2 | 2 | 4 | S |
| 65 | WH542 | Yr64 | Yr16, Yr18, Yr30, Yr59 | 1 | 4 | 5 | R |
| 66 | WH1105 | Yr2, Yr10 | Yr16, Yr30 | 2 | 2 | 4 | R |
| 67 | HD2967 | Yr2, Yr7, Yr24, Yr64 | Yr30 | 4 | 1 | 5 | S |
| 68 | Kharchia | Yr7, Yr24 | Yr16, Yr30 | 2 | 2 | 4 | S |
3.3 Molecular identification of Yr2
SSR marker Wmc364 linked to the yellow rust resistance gene is located on chromosome 7B at the distance of 5.6 cM [43]. Wmc364 amplified two alleles (+200 bp/–190 bp) for distinguishing gene positive and negative genotypes. A total of 31 genotypes (45%) amplified 200 bp alleles whereas 16 genotypes (23.5%) amplified both the alleles (HI 8759, HI 8774, HPPAU 05, WB 2, DBW 179, DBW 216, DBW 217, DBW 219, DDK 1050, DDK 1051, MACS 5044, RKD 292, DBW 220, HS 580, VL 3012 and WH 1310). Twenty-one genotypes (30.8%) amplified 190 bp allele (HS 626, RKS 283, TL 3006, TL 3007, TL 3008, TL 3009, UAS 459, Infector, HD 3209, HI 1605, K 1317, AKAW 4842, NW 6046, PDW 344, VL 3002, VL 3011, WH 1216, Lassik, WH 542, Kharchia and WH711). On the basis of screening at genotypic level, 47 genotypes (69.1%) were postulated to carry resistant allele for Yr2 gene. Since the distance between marker and gene is far, it is not recommended for marker assisted breeding.
3.4 Molecular identification of Yr5
Yellow rust resistance gene Yr5, originally derived from Triticum spelta var album [44] is a race-specific R-gene effective at both seedling and all plant growth stages and located on the chromosome 2BL [45]. Four microsatellite markers Xgwm501, Wmc175, Barc349, Barc167 [46] and two STS markers S19M93-100 [47], STS7/8 [48] linked with yellow rust resistance gene Yr5 were used to confirm and evaluate the diagnostic potential of these markers in wheat genotypes. Microsatellite marker Wmc175 and Barc167 failed to yield any amplicons whereas S19M93-140 located at 0.54 cM from Yr5 amplified one 100 bp allele. Screening with S19M93 marker produced the expected 100 bp band associated with Yr5 gene in 30 genotypes (44.1%), while the other 38 wheat genotypes (55.8%) failed to amplify the gene. Another marker STS7/8 is proximally located 0.3 cM from the Yr5 gene and amplified 500 bp bands in 23 genotypes (33.8%) with Yr5 gene. Dominant SSR marker Barc349 is located 0.4 cM distally [48] and amplified 5 alleles (100 bp, 105 bp, 120 bp, 140 bp, 220 bp). Product size 105 bp that is linked with presence of Yr5 gene was observed in 32 lines (47%). SSR marker Xgwm501 is located 10.5–13.3 cM from Yr5 gene [49] and amplified 3 alleles (100 bp, 150 bp, 176 bp). Thirty-one genotypes (45%) produced the expected 176 bp band which indicated that they may contain Yr5. However, the genetic distance between the markers Xgwm501 and Yr5 is too far to be used in marker-assisted selection. On the basis of proximal and distal distance of Yr5 linked markers, 12 genotypes (17.64%) (HS 626, TL 3007, HD 3209, K 1317, DDK 1051, PDW 344, WH 1184, HS 510, VL 3010, VL 3011, VL 3012 and WH 1181) were identified with Yr5 gene.
3.5 Molecular identification of Yr7
Yr7 was first identified in wheat cultivar ‘Lee’ (Triticum turgidum (lumillo durum) [50] and mapped on chromosome 2BL [51]. Yr7 is allelic to Yr5 and linked with Sr9 g [52]. Microsatellite marker Xgwm526 linked with Yr7 locus at 2.3 cM was used to detect the presence of Yr7. Thirty-nine genotypes (57.4%) showed 145 bp fragment associated with the presence of Yr7. The remaining 29 wheat genotypes did not show 145 bp products, indicating likely absence of Yr7. SSR marker CFD77 amplified a 220 bp allele in all the genotypes indicating non-diagnostic behaviour of that marker. Overall 36 genotypes (52.9%) were postulated to harbour the Yr7 gene.
3.6 Molecular identification of Yr9
Yr9, located on the short arm of rye chromosome 1R, was transferred to wheat through chromosomal translocation of 1BL.1RS. Four markers STSSr31[53], Xgwm582, IB267 and Xwgp8 were chosen to amplify Yr9 gene. STS marker Sr31 and RGAP marker Xwgp8 failed to give amplification reactions. Xgwm582 is present on chromosome 1BL at 3.7 cM distance of Yr9 and amplified 150 bp product size in 23 genotypes (33.9%) carrying Yr9 gene [54]. IB267 marker is completely linked at 0 cM [55] and produced Yr9 specific 267 bp bands in 2 genotypes (PBW 723 and HPPAU 05). Based on 2 sets of primer pairs, PBW 723 and HPPAU 05 genotypes amplified both of the primers and were postulated to carry Yr9.
3.7 Molecular identification of Yr10
Dominant gene, Yr10 was isolated from Moro [21] and located on chromosome 1BS, 2 cM apart from Rg1 locus that confers brown glume colour [56] and 5 cM from locus Gli-1B [57]. Microsatellite marker Xpsp3000 located 1.3 cM proximal to Yr10 [58] was used to determine the presence or absence of the gene in wheat genotypes. On screening with marker Xpsp3000, a varied range of allelic variation (220 bp, 240 bp, 260 bp and 285 bp) at Yr10 locus was observed. Thirty-four genotypes produced 260 bp fragment associated with the presence of Yr10 allele [59], 16 genotypes showed 285 bp fragment specific for Yrvav allele [60], 48 genotypes amplified 240 bp fragment specific for susceptible allele. Seven genotypes showed amplification of both 285 and 260 bp allele (Yr10 + Yrvav) (K 1317, DBW 216, PDW 344, WH 1184, HS 580, VL 3011, HPBW 02), 15 genotypes amplified both 260 bp and 240 bp alleles (HI 8759, HI 8774, PBW 756, RKD 283, TL 3007, TL 3008, TL 3009, WH 1214, GW 477, UAS 459, MACS 5046, NW 6046, HPPAU 10, WH 542, WH 1105). Five genotypes amplified all four alleles (HPW 423, DBW 179, PBW 621, VL 4001 and HPPAU 08) and 31 genotypes amplified a 220 bp allele in combination with other alleles indicating the presence of a novel allele at Yr10 loci. One Genotype, WB2 was identified carrying 260 bp allele. Both 260 bp and 285 bp bands are associated with the presence of resistance gene. On this basis, 34 genotypes (50.0%) carried Yr10 gene.
3.8 Molecular identification of Yr15
Dominant gene Yr15, derived from Triticum dicoccoides, is located on chromosome 1BS [61]. Murphy et al. [48] showed that Xbarc8 and Xgwm413 were closely linked with Yr15. The Yr15 gene was mapped to a 6.4 cM interval flanked by marker Barc8, located 3.9 cM to the distal side, and by Xgwm413 and Xgwm273 located 2.5 cM and 2.1 cM to the proximal side. Six markers, Xgwm11, Xgwm18, Xgwm 33, Xgwm273, Barc8 and Xgwm413 linked with Yr15 were used for validation purpose. SSR marker Barc8, produced 2 alleles (260 bp and 280 bp), respectively. Thirty genotypes (44.1%) amplified 260 bp bands associated with Yr15 gene while 37 genotypes (54.4%) amplified 280 bp without Yr15 gene. SSR locus Xgwm 413 located 3.5 cM proximally produced 3 alleles (90 bp, 95 bp and 100 bp) among wheat genotypes used in this study. A huge variation in allelic profile of Xgwm413 was observed. Forty-three genotypes amplified 90 bp specific alleles, 64 genotypes amplified 95 bp alleles, 36 genotypes amplified 100 bp alleles, 17 genotypes produced both the 95 bp and 100 bp alleles, 25 genotypes amplified 90 bp and 95 bp alleles and 17 genotypes amplified all the three alleles (90 bp, 95 bp and 100 bp). Xgwm273 marker is present 0.4 cM from Yr15 and amplified 7 alleles (156 bp, 165 bp, 180 bp, 200 bp, 350 bp, 400 bp and 500 bp), respectively. Sixteen genotypes (23.5%) produced 156 bp bands specific for Yr15 [62]. Xgwm11 and Xgwm18 failed to give amplifications. Xgwm33 produced inconclusive results. On the basis of proximal and distal distance of linked markers 25 genotypes (36.7%) (HI 8774, HPPAU 05, HS 628, PBW 725, TL 3007, HD 3209, AKAW 4842, DBW 179, DBW 216, DBW 217, DBW 219, DDK 1050, GW 477, MACS 5044, MACS 5046, PBW 621, RKD 292, VL 4001, WH 1215, DBW 220, HPBW 02, HPPAU 10, HPW 424, HS 627 and VL 3010) were predicted with Yr15.
3.9 Molecular identification of Yr16
Yr16 was originally originated from the French cultivar Capelle-Desprez and mapped on chromosome 2DS. SSR, Xgwm102 corresponding to the QTLs (QYr.ufs-2D) is reported to be linked with gene Yr16 and was used to screen the wheat genotypes [63]. SSR marker, Xgwm249 linked to Yr16 gene showed polymorphism in present investigation with product size ranging from 120 to 160 bp. Since, no specific product size information is available for this marker. Screening results could not be associated with Yr16 gene. Marker Xgwm102 amplified three alleles 150 bp, 155 bp and 200 bp, respectively. Twenty-two wheat genotypes (32.3%) produced the 155 bp and 200 bp allele confirming the likely presence of this gene [63].
3.10 Molecular identification of Yr17
Three rust resistance genes i.e. Yr17, Lr37 and Sr38 were translocated into the short arm of wheat chromosome 2A of bread wheat from 2NS chromosome of Triticum ventricosum [64]. The presence of Yr17 gene in Indian wheat genotypes was investigated with the primers VENTRIUP/LN2 [24]. All the wheat genotypes amplified a 259 bp fragment specific for Yr17 gene, indicating either the presence of Yr17 in all the genotypes or the non-diagnosing behaviour of this marker in the Indian wheat background. Therefore, further confirmation of Yr17 was done with the help of primers URIC/LN2 which amplifies the two fragments of 285 bp (N-allele) and 275 bp (A-allele). Subsequent digestion of the undigested PCR products with restriction enzyme DpnII yields the 275 bp and the 166, 109 bp fragments corresponding to the presence and absence of gene. None of the wheat genotypes, other than the positive control Lassik gave positive restriction digestion results for Yr17 gene, confirming the likely absence of Yr17 gene in this panel of Indian wheat genotypes.
3.11 Molecular identification of Yr18
The presence of yellow rust resistance gene Yr18 was assayed using STS marker csLV34, gene specific marker cssfr2 and SSR marker Xgwm295. The STS marker csLV34 located 0.4 cM distal to Yr18 [65] produced two allelic variants i.e. csLV34b and csLV34ba. The 150-bp band (csLV34b) and 229-bp band (csLV34ba) indicated the presence and absence of Yr18 gene, respectively. Twenty-four genotypes (35.3%) amplified 150 bp fragments, suggesting the presence of Yr18-associated allele. Forty-four genotypes (64.7%) produced 229 bp band associated with csLV34a allele indicating absence of Yr18 gene. The gene specific marker cssfr2 produced 517 bp products with Yr18. Thirty-five genotypes (51.5%) amplified cssfr2 marker. A total of 42 genotypes (61.8%) showed the association with Yr18 gene by using any of the markers while 20 genotypes (29.4%) (HI 8759, HI 8774, HPPAU 05, HPW 423, HPW 433, HS 623, TL 3006, TL 3007, MACS 5044, MACS 5046, NW 6094, HPBW 02, HPW 424, HS 580, VL 1009, VL 3002, VL 3010, VL 3011, Lassik and WH 542) were positive with both the markers. SSR marker Xgwm295 produced variable results and could not be linked with the presence of gene.
3.12 Molecular identification of Yr24/26
Yr24 confers ASR to yellow rust and was originally identified in T. turgidum var. durum accession K733. Yellow rust resistance gene Yr26, carried by T. turgidum durum line and was located on chromosome 1B [66]. Yr26 and Yr24 are considered to be identical genes due to their disease reaction against rust isolates [67]. Two markers Barc181 and Barc187, were used to detect the presence/absence of Yr24/Yr26 genes. The SSR marker Barc187 linked with Yr24 at the distance of 2.3 cM [68], amplified three alleles; 200 bp, 220 bp and 225 bp, respectively. Forty genotypes (58.9%) amplified 200 bp indicating the likely presence of Yr24, whereas 31 genotypes (45.6%) produced 220 bp and 225 bp alleles, without Yr24. Microsatellite marker Xbarc181 is 6.7 cM distal to Yr26 [68] and produced 2 alleles 180 bp and 200 bp, respectively. Thirty-eight genotypes (69.2%) produced 180 bp allele associated with Yr26 gene while 34 genotypes amplified the 200 bp specific allele. Four genotypes (HS623, HS626, DDK1051 and DBW220) amplified both the alleles. Combined results with both the markers exhibited the presence of Yr24/Yr26 in 19 genotypes (27.9%) i.e. HPPAU 05, HPW 423, HS 622, PBW 725, TL 3009, HI 1606, K 1317, AKAW 4842, DBW 217, DBW 219, DDK 1050, PBW 621, WH 1215, HPBW 02, HPPAU 08, HPPAU 10, HS 627, NW 6046 and VL 1009.
3.13 Molecular identification of YrCH52
Xgwm498 located at 1.6 cM produced 160 bp associated with YrCH52 in 25 genotypes (36.7%), EST based marker CON-6 (0 cM) and STS marker CYS-5 (0.5 cM) developed from resistance gene analogues [69] did not produce any results.
3.14 Molecular identification of Yr25
Xgwm6 linked with Yr25 gene produced monomorphic bands of 150 bp in all the genotypes indicating non-diagnosing nature of the primer.
3.15 Molecular identification of Yr27
Yr27, originated from the cultivar McMurachy and is located on chromosome 2BS [70]. The chromosomal region of Yr27 harbours Lr13, Lr23 and Lr31 rust resistance genes. SSR marker Xgwm630 is located 10 cM from Lr13 (Seyfarth et al., 1999). Since, Yr27 is linked with Lr13; Xgwm630 was used to check the abundance of Yr27 gene in wheat genotypes [71]. Xgwm630 primer amplified a 124 bp allele in all the genotypes marking either the 100% abundance of Yr27 or its absence. Nevertheless, the genetic distance (10 cM) is too large for use of Xgwm630 for MAS. Yr27 is an all plant stage resistant gene that shows the durability in combination with Lr34 and other resistance genes [72]; however, as a single gene Yr27, is no longer effective in most wheat growing areas (McIntosh et al., 1995) [73].
3.16 Molecular identification of Yr29
Yr29 was originally identified in cultivar Pavon 76 and was assigned to chromosome 1BL [74]. Yr29 confers moderate level slow-rusting APR to yellow rust. Yr29 gene is closely linked to Lr46 for leaf rust resistance. Wmc44 flanks Yr29 3.6 cM proximally [75] and amplified 8 alleles (205 bp, 210 bp, 220 bp, 240 bp, 260 bp, 280 bp, 300 and 310 bp), respectively. Twenty-six genotypes (38.2%) specific as predicted for (260 bp) with Yr29 and 49 lines (72%) without Yr29 were identified. SSR Xgwm 259 gave inconclusive results.
3.17 Molecular identification of Yr30
Yr30 is a minor APR gene introgressed from Yaroslav emmer wheat into bread wheat along with the stem rust resistance gene Sr2 [76] and is located on chromosome 3BS [77]. Yr30 is closely linked with Sr2 for stem rust resistance and Lr27 for leaf rust resistance [78]. SSR marker Xgwm533 (+120 bp) has been used in MAS for incorporating the Sr2 gene into wheat cultivars (Spielmeyer et al., 2003) hence Xgwm533 can be potentially used to select Yr30 gene containing genotypes. Screening with Xgwm533 amplified 5 alleles 100 bp, 120 bp, 130 bp, 140 bp and 150 bp, respectively. Forty-six genotypes (67.6%) produced 120 bp bands specific for Yr30 while 10 genotypes (14.7%) failed to yield any amplicons. CAPS marker csSr2 failed to amplify any of the genotypes.
3.18 Molecular identification of Yr32
Yr32 was originally detected from cultivar Carstens V [79] and is located on chromosome 4D. SSR marker WMC198 is closely linked to Yr32 at 0.2 cM [80] and amplified two alleles 180 bp and 200 bp, respectively. Six genotypes (8.8%) (HD 3209, AKAW 3842, DBW 216, DBW 219, RKD 292 and VL 1009) produced bands associated with Yr32 while the remaining genotypes failed to amplify the gene.
3.19 Molecular identification of Yr35
Yr35 confers effective all-stage resistance. It was derived from T. dicoccoides and was located on chromosome 6BS [81]. SSR marker cfd1 located at 4.1 cM from Yr35 and 3.0 cM to Lr53 gene didn’t amplify in any of the genotypes indicating absence of Yr35 gene in all genotypes.
3.20 Molecular identification of Yr36
Yr36 is derived from T. turgidum ssp. dicoccoides accession FA15-3 and confers high level of HTAP resistance. It is located on chromosome 6B. Three primers, ASA at 0.3 cM [82], SSR Barc101 [82] at 2 cM and WKS1 (gene based) [83] linked with Yr36 were used to screen the wheat genotypes. ASA primer amplified 1.6 kb product size in 3 genotypes (4.5%) i.e. HPPAU 05, AKAW 3842 & Lassik. Barc101 amplified 5 alleles 116 bp, 124 bp, 138 bp, 160 bp and 165 bp respectively. Twenty-seven genotypes (39.7%) produced a 165 bp band associated with Yr36. Remaining genotypes showed absence of this allele. WKS1 is HTAP gene-based marker and amplified 128 bp product sizes in two genotypes (HPPAU 05 and Lassik).
3.21 Molecular identification of Yr40
Yr40 originated from ovate goat grass, Ae. geniculata Roth (syn. Ae. ovata L.) is located on chromosome 4D and confers effective ASR to yellow rust. Yr40 and Lr57 was either closely linked genes or they are the same gene with pleiotropic effect [84]. SSR Xfbb276 amplified 1 kb and 2 kb alleles in 20 lines (29.4%) indicating likely presence of Yr40.
3.22 Molecular identification of Yr46
Yr46 confers APR to yellow rust and is located in the middle of chromosome 4DL. Cfd71 and Cfd23 markers are associated with Lr47 gene [85]. Cfd71 amplified 3 alleles 148 bp (+), 150 bp and 152 bp while Cfd23 amplified two alleles of 214 bp and 211 bp in 20 genotypes (29.4%). On the basis of both primers 13 genotypes (19.1%) (K 1317, DBW 219, MACS 5044, WH 1215, DBW 220, HPBW 02, HS 580, VL 1009, UP 2954, UP 2955, VL 3002, VL 3011 and VL 3012) identified with Yr46 gene.
3.23 Molecular identification of Yr47
Yr47 (YrW1) originated from Iranian wheat landraces and was located on chromosome 5BS [86]. SSR marker cfb309, estimated to be about 8–12 cM from Yr47. Yr47 is linked with leaf rust resistance gene Lr52 with a genetic distance of 3–6 cM [86]. Cfb309 amplified 2 alleles of 350 bp and 600 bp. Allele 600 bp is associated with the presence of Yr47 and was found in 6 genotypes (8.8%) while 15 genotypes (22.05%) produced both alleles indicating heterozygosity of the gene. Four genotypes (5.8%) produced 350 bp allele confirming absence of this gene. Remaining genotypes didn’t amplify any product. Twenty-one genotypes (30.9%) are presumed to include the Yr47 gene.
3.24 Molecular identification of Yr48
Yr48, derived from wheat genotype PI610750 is a QTL conferring APR to yellow rust and located on chromosome 5AL [87]. Marker-assisted breeding programs uses closely linked markers SNF-A2 (0.18 cM proximal of Yr48), BE495011 (0.09 cM proximal of Yr48) and cfa2149 (0.06 cM distal of Yr48) for selection of Yr48 gene. SSR cfa2149 produced 2 alleles of 225 bp and 250 bp. Twenty-two Genotypes (32.4%) showed both 225 bp and 250 bp alleles, 2 genotypes (2.9%) showed 225 bp, 2 genotypes (2.9%) showed 250 bp alleles. SNF-A2 amplified 3 alleles of 150 bp, 180 bp and 220 bp. Two genotypes (2.9%) (TL 3009, UAS 459) produced 150 bp alleles and were associated with Yr48. Two genotypes (2.9%) gave 180 bp and 220 bp alleles, five genotypes produced 180 bp allele and eight genotypes produced 220 bp allele. BE495011 amplified one allele of 220 bp in 18 genotypes (26.5%) confirming presence of this gene. Overall 32 genotypes (48.5%) confirmed positive with all these markers. Combining proximal and distal position, 12 genotypes (17.6%). HI 8759, HI 8774, HPW 433, UAS 459, NW 6094, PBW 621, PDW 344, WH 1184, UP 2955, WH 1310 and Lassik were postulated to carry Yr48.
3.25 Molecular identification of Yr51
Yr51 is recessive in nature and confers ASR to yellow rust. Yr51 is delimited by markers owm45F3R3 at 1.2 cM proximal and sun104 at 2.5 cM distal, respectively, locating Yr51 on the long arm of chromosome 4B. Since, Yr51 is not present in modern wheat genotypes, positive validation was not feasible. Marker sun104 amplifies 225 bp in resistant lines and a null in susceptible lines. Negative validation using sun104 produced null allele in all 68 genotypes confirming absence of Yr51 gene. Marker owm45F3R3, amplified in 10 genotypes (14.8%). Since, no earlier data on this marker is available this marker was considered unsuitable for diagnostic purpose. Therefore, sun104 can be used for marker-assisted selection of Yr51 in wheat genotypes lacking the resistance linked 225 bp allele. Marker assisted detection for Yr51 revealed the absence of this gene in Indian wheat genotypes. Similar results were obtained by Randhawa et al. [27].
3.26 Molecular identification of Yr59/YrZH84
Yr59, identified from an Iraqi spring wheat landrace, PI 178759, codes for HTAP resistance and is located on chromosome 7BL [28]. SSR locus Xbarc32, located proximal to Yr59 (< 2.1 cM) produces a 165 bp product in the resistant line and a 175 bp product in the susceptible line. On screening with this primer, 4 alleles were amplified 165 bp (+), 175, 190 bp and 250 bp, respectively. Eleven genotypes (16.2%) were found to contain this gene, while the remaining genotypes did not amplify the 165 bp allele associated with Yr59. On the distal side Xwmc 557 is located (> 2.2 cM). Screening with this primer amplified 2 alleles, 315 bp and 500 bp. Thirteen genotypes showed presence of this gene indicating 19.2% polymorphism rate. Over all 17, genotypes (25%) amplified either of the primers while 4 genotypes (HI 8759, DDK 1050, VL 1009 and UP 2954) showed amplification with both of these primer pairs.
3.27 Molecular identification of Yr60
Yr60 (YrLalb) resistance gene is developed from the Mexican wheat line ‘Almop’ (Wiliam et al., 2003) [88] and located on the distal end of chromosome arm 4AL. This gene confers moderate resistance at both the seedling and the adult-plant stages. SSR locus wmc776 located < 0.6 cM away amplified 3 alleles 150 bp, 160 bp and 170 bp respectively. Fourteen genotypes (20.6%) amplified this gene with 150 bp, 160 bp and 170 bp respectively. Wmc313 is located 0.6 cM distal to Yr60 and amplified 180 bp and 200 bp alleles in 20 genotypes (29.3%) whereas wmc219 which is also located 0.6 cM to Yr60 amplified 200 bp and 220 bp alleles in 16 genotypes (23.6%). Remaining genotypes didn’t amplify any product confirming absence of this gene. Combined results using all the markers predicted Yr60 in HI 8774 (d).
3.28 Molecular identification of Yr64
Yr64 confers ASR to yellow rust and is located on chromosome 1BS. SSR Xgwm413 is linked to Yr64 gene distally at 3.5cM [30]. Screening of wheat genotypes with Xgwm413 led to the amplification of 95 bp allele in 42 genotypes, predicting its likely presence in the Wheat breeding lines.
3.29 Molecular identification of YrMor
S26M47 amplified 250 bp product size associated with the presence of the gene in 10 genotypes (14.8%) i.e. HPPAU 05, HPW 433, PBW 760, TL 3000, DBW 316, VL 4001, HPPAU 08, WH 1216 & HD 3171.
PM47 marker specific for YrHua gene, Xgwm410 specific for YrCN19/Yr41, WMC631 specific for YrExp1 gene and WMC441 associated with YrSPp gene did not produce any result confirming either the absence of this gene in wheat genotypes or non-diagnostic behaviour of markers used for the screening purposes. RGAP markers linked with Yr9, Yr45 also didn’t yield satisfactory results may be due to non-specificity of the markers in diverse wheat background. Electrophoretograms of various Yr genes are given in Fig. 1.
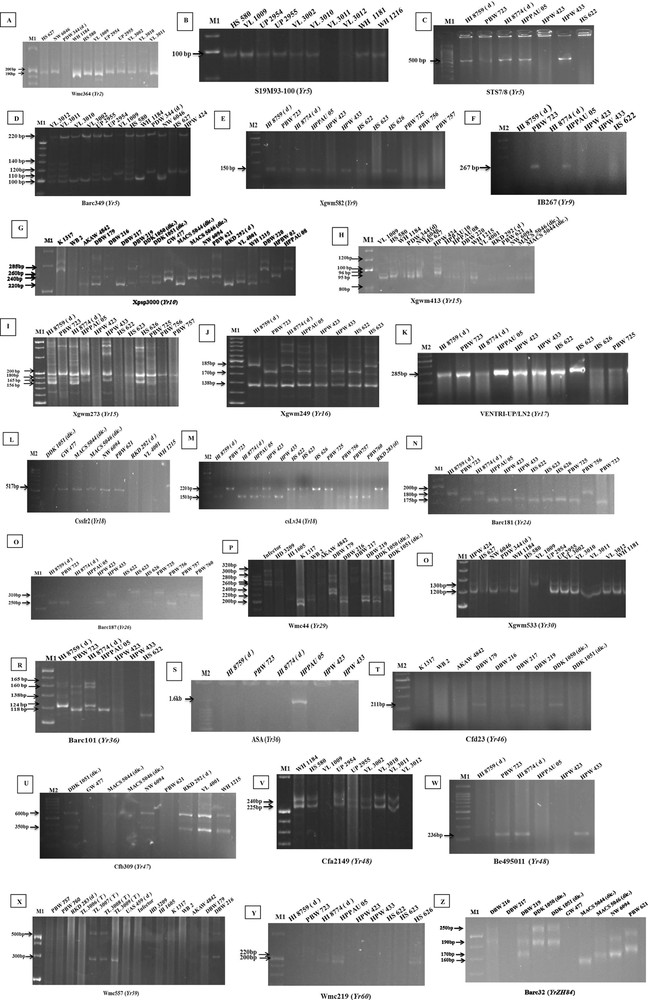
Electrophoretogram of the primers for different Yr resistance genes. The corresponding Primers of Yr2, Yr5, Yr9, Yr10, Yr15, Yr17, Yr18, Yr24, Yr26, Yr29, Yr30, Yr36, Yr46, Yr47, Yr48, Yr59, Yr60, and YrZH84 were used to screen the elite wheat lines and varieties (A–Z). Although no control accession was used, the postulated bands were easy to be distinguished according to previously reported base pairs of the PCR product. The presence or absence of gene is marked with an arrow. Note: M1, 20 bp DNA Ladder (Fermentas life sciences); M2: 100 bp Ladder (G Biosciences).
4 Discussion
The severe yellow rust problem on the predominant wheat varieties of India is due to the excessive dependence on seedling resistance genes Yr2, Yr9 and Yr27 in isolation from other APR genes and absence of major avirulent (Yr10, Yr15 & Yr24/Yr26) genes in the adopted varieties. Rapid evolving and virulent races of Pst have overcome the resistance of these major single genes deployed in widely cultivated varieties with the passage of time [89]. The breakdown of resistance has alarmed wheat breeders in the need for broadening the genetic base of future Indian wheat varieties by incorporating multiple yellow rust resistance genes. Identification of novel genes for yellow rust resistance can help breeders to efficiently and accurately incorporate those genes into breeding material by MAS to reduce the disease incidences. Deployment of specific gene combinations provides durable and improved resistance versus using single genes because single specific gene is subject to become susceptible due to genetic shifts in the pathogen [90]. Therefore, 70 closely linked markers specific for 35 Yr genes were used to detect the likely presence of Yr genes among the wheat genotypes and evaluate their contribution to the current status of Pst resistance. Most of the genes were amplified by using 1–5 linked markers for better accuracy of results. The combined results of all the markers were used for confirming the presence of genes. However, those markers which are gene based have better accuracy and can be considered alone for evaluation of resistance genes in wheat breeding programmes. Our molecular screening results correlated with the published reports for the size of expected PCR product and provides useful information for choosing specific resistant genes and an opportunity to pyramid the resistant genes against Pst pathotypes in a single wheat genotype. Out of 35 Yr genes, 25 genes amplified in wheat genotypes under study. The distribution of 25 Yr resistance genes among 68 wheat genotypes is illustrated in Fig. 2.
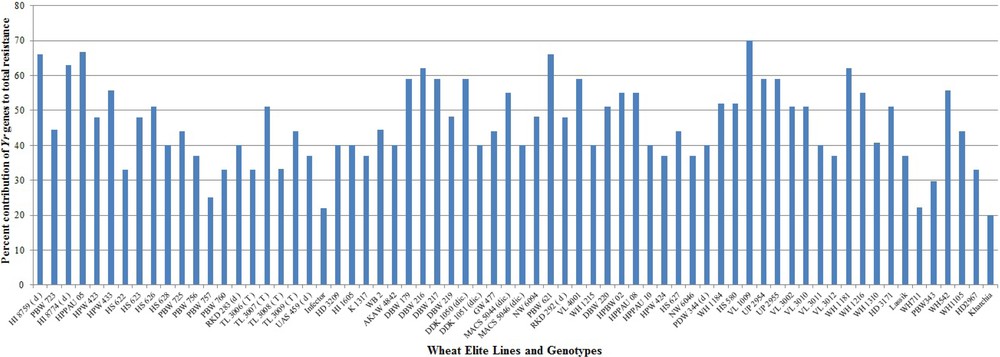
Percent contribution of Yr genes to the total resistance in wheat genotypes.
Two genotypes (VL 1009 & VL 3002) were identified with maximum 15 Yr genes followed by 14 genes in VL 3010 and HI 8759, respectively. Both of these genotypes exhibited resistance response at field level. Most 11 seedling resistance genes were identified in RKD 292 followed by 10 genes in DBW 216, WH 1184 and VL 3002 while most six APR genes were identified in HI 8759 and VL 3009 followed by five genes in, Lassik respectively. Even though RKD292 contained the major Yr genes (Yr10, Yr15, Yr24 & Yr26) and none of the APR gene, it still exhibited resistance response at field level concluding the major effects of these genes. Lowest numbers of two Yr genes were detected in genotypes PBW 757 and WH 711 susceptible genotypes. No major Yr genes were identified in susceptible genotypes. None of the RS/ASR genes were identified in genotype WH 711 while PBW 757 had one gene, followed by two genes in TL 3006, WH 1105, Kharchia and PBW 343 genotypes, respectively. Genotypes (HI 1605 and RKD 292) were identified with none of the APR genes but presence of Yr10, Yr24 & Yr26 imparted resistance response at field testing.
The genotypes carrying multiple Yr genes might be useful for pyramiding Pst resistance sources into commercial varieties [35]. The ratio of RS and APR genes to the total resistance among wheat genotypes is illustrated in Fig. 3. The higher frequency of RS genes over APR genes in Indian genotypes indicates an erosion of APR genes resulting from preferred use of dominant RS and ASR genes which were easier to select and breed into existing crop cultivars in Indian breeding programs. Genes Yr26 (69.2%), Yr2 (69.1%), Yr64 (61.7%), Yr24 (58.9%), Yr7 (52.9%), Yr10 (50%) and Yr 48 (48.5%) showed high frequency among our panel of wheat genotypes, while Yr9 (2.94%), Yr36 (2.94%), Yr60 (1.47%) and Yr32 (8.8%) were least frequent in wheat genotypes (Fig. 4). Similar frequency of Yr genes among genotypes was observed in previous studies [18,91]. Zheng et al. [16] also studied the molecular characterisation of 330 leading wheat cultivars and 164 advanced breeding lines in China and identified Yr9, Yr17, Yr18 and Yr26 in 134 (29.4%), 45 (9.1%), 10 (2%) and 15 (3%) entries, respectively.
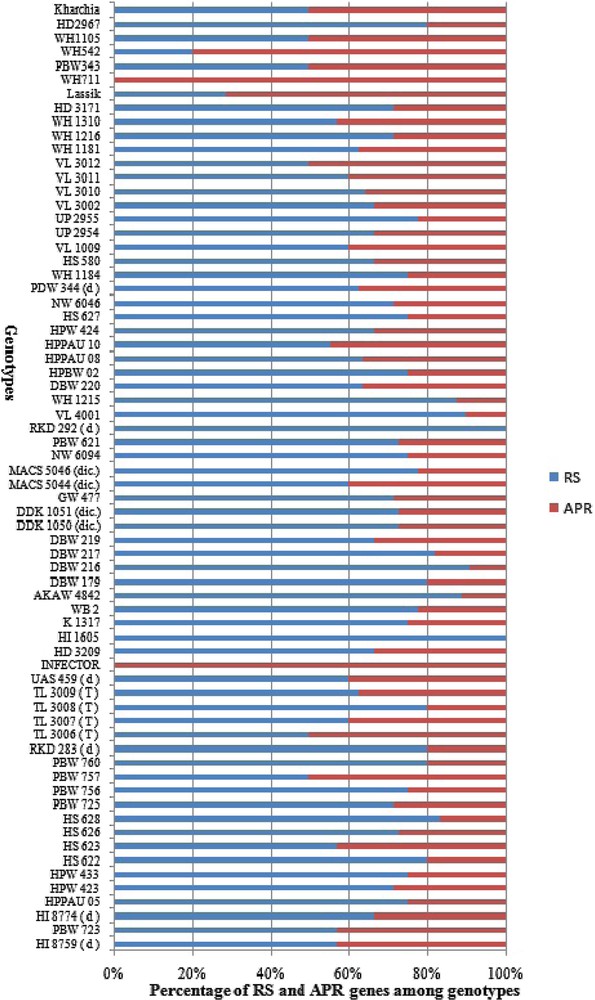
Contribution of all-stage resistance genes (RS) and adult-plant resistance (APR) genes in wheat genotypes.
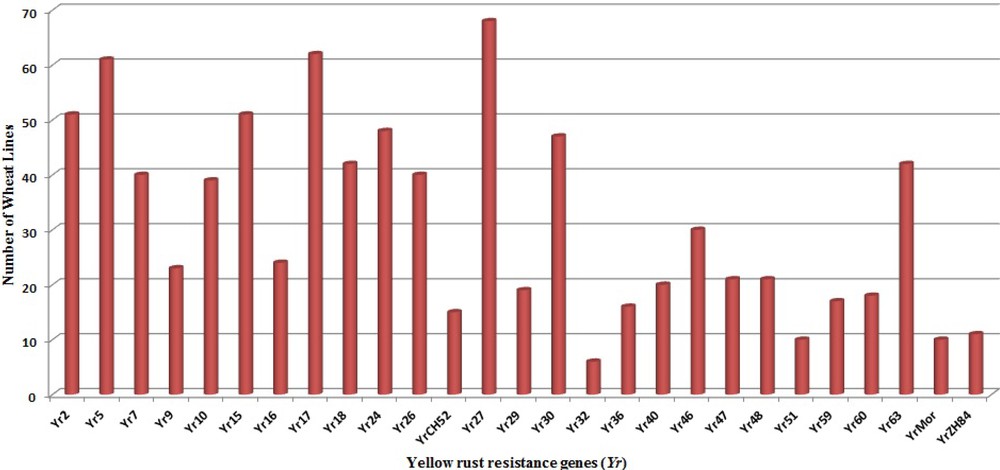
Abundance of individual Yr genes in wheat genotypes.
Yr5 is found to be effective against all rust virulent races in North America [7,20,92], Iran, China [7,93], India and Turkey [94]. Yr5 gene is still resistant to major races in India and has potential to become an effective source of yellow rust resistance if deployed in combination with other R-genes. On screening with S19M93 marker specific for Yr5 gene, polymorphic rate of 44.1% was observed among wheat genotypes. Ullah et al. [32] obtained 89% polymorphic rate of Yr5 gene in 99 Pakistan wheat lines on screening with S19M93 molecular marker. Our findings for Yr5 gene were also in accordance with the screening results of Iqbal et al. [33]. On the basis of combined results of the entire Yr5 markers, all of the Yr5 gene containing twelve genotypes (TL3007, K1317, WB2, NW6046, WH1184, HS580, VL3010, VL3011, VL3012, WH1181, HD3209, DDK1051 & VL3002) showed fairly good to moderate resistance response at field testing. A total of 52.9% genotypes showed the presence of Yr7 gene. Yr7 is known to be allelic to Yr5 and linked with stem rust Sr9 g gene [52]. These genotypes can be further tested against stem rust pathotypes to be used as donor in stem rust breeding programmes. Yr10 gene has also been reported to be effective against all races in India, Pakistan, China, Iran and USA [95]. Bariana et al. [60] reported two alleles for Yr10 i.e. Yr10 and Yrvav and described that varieties with Yr10 amplify a 258–260 bps fragment, 285 bp with Yrvav allele and 240 bp for lacking the Yr10 gene. In our study, a 220 bp allele was also identified among the thirty-one wheat genotypes. Yaniv et al. [96] also reported the presence of 220 bp in Pakistani wheat varieties and linked it to the Yr10 resistance gene. Totally, 50% genotypes exhibited the presence of Yr10 resistant allele. Most of the Yr10 alleles co-segregated together indicating the heterozygosity of Yr10 locus. Similar, heterozygous pattern was reported in 28% varieties using Xpsp3000 marker by Talha et al. [97]. However, close genetic distances have been reported to exist between reported marker gene combinations. Though the chance of recombination is very low but could not be neglected. Therefore, the allelic variability among genotypes also emphasizes the need of gene-based markers for efficient selection in marker assisted programmes. A close association between Xpsp3000 marker and Gli-B1 gene has also been reported [57]. Gli-B1 encodes a wheat storage protein which improves plant resistance to abiotic stresses. This association of Yr10vav and Yr10 with specific alleles of Gli-B1 and Xpsp3000 can be useful in marker-assisted selection and gene pyramiding. Marker validation depends on effective marker/trait linkage. To further validate the presence of Yr5 and Yr10, an independent F2 population can be developed from cross between positive parents and a highly susceptible line which can be tested for phenotypic segregation in field and genotypic segregation by proposed markers for respective genes.
The Lr34/Yr18 gene for rust resistance has been used in agriculture for centuries. In contrast to many other resistance sources against leaf rust and stripe rust, it has remained effective and no virulence has been reported. In our study, the frequency of Yr18 allele (29.4%) seemed to be comparatively higher in Indian breeding lines than the previously reported studies. All the genotypes exhibited resistant response at field testing. One genotype, MACS 5046 showed moderate response. Wu et al. [98] found that many Chinese wheat landraces that were predicted to possess the Yr18 resistance allele by marker assays were highly susceptible to yellow rust in the field. This indicated that such landraces either contained susceptible allelic variations at the Yr18 locus or a suppressor of gene function. Yang et al. [99] identified Lr34/Yr18 genes in 231 Chinese wheat cultivars and 422 landraces using the STS marker csLV34 with a frequency of 6.1% and 89.6% respectively. Similarly, Alma et al. [100] characterized elite wheat germplasm from Central Asia using molecular markers linked to the Lr34/Yr18 and found a 16.7% frequency of the csLV34b-allele. As this gene complex provides durable adult-plant resistance, its deployment should be increased in future wheat varieties of India so that their genetic base can be broadened against the continually evolving new races of P. striiformis tritici.
Yr24, Yr26 and YrCH42 are considered to be identical genes due to their similar infection types against rust isolates [67]. Combined results with markers exhibited the presence of Yr24/Yr26/YrCH52 in 19 genotypes (27.9%). All these genotypes exhibited significant lower ACI values. Yr24/Yr26/YrCH52 imparts race specific seedling resistance therefore, in wheat breeding programs; it should be used in combination with other major or minor resistance genes. Yang et al. [101] suggested the importance of gene pyramiding in improvement of durability of rust resistance.
Marker assisted detection for Yr51 using negative validation by sun104 marker revealed the absence of this gene in Indian wheat genotypes. Similar results were obtained by Randhawa et al. [27]. The genetic association of yellow rust resistance with other traits also allows indirect or direct selection for resistance in breeding. Examples include Yr18/Lr34/Sr34, Yr9/Lr26/Sr31, Yr17/Lr37/Sr38, Yr18/Lr34/Pm38, Yr29/Lr46, Yr40/Lr57 and Yr46/Lr67/Sr55. Yr60 gene was identified in one genotype HI 8774 (d). Apart from Yr60 gene, Wmc219 and Wmc313 markers are also linked with leaf rust resistance gene (Lr28) and Septoria tritici blotch resistance gene (Stb7) at 5 cM [102] and 0.5–1.1 cM, respectively [103]. Genotype, HI 8774 can be tested at field level for the presence of leaf rust and septoria blotch resistance for durable rust breeding programmes.
Lines with these genes can be utilised alone or in combination with other ASR and APR genes containing lines. This type of resistance has been used in CIMMYT programs for improvement of leaf rust resistance) [104]. Some genotypes showed good frequency of APR genes such as Yr18 (29.4%) loci which also coincides with leaf rust, stem rust and powdery mildew disease resistance locus, Yr 30 (67.6%), Yr 59 (19.2%) and Yr46 (19.1%). Disease severity response was recorded from resistant to moderate susceptible (0–10 MS) among the genotypes containing these APR genes. Yr30 is a minor APR gene and is closely linked with Sr2 for stem rust resistance and Lr27 for leaf rust resistance [78]. Forty-six genotypes (67.6%) postulated with Yr30 showed effective resistance in the field testing. Since Yr30 is linked with Sr2, it can be effective source in stem rust breeding programmes. Yr46 is a pleiotropic gene and confers APR to yellow rust. It is also known to confer resistance to stem rust (Sr55), leaf rust (Lr67), powdery mildew (Pm46) and also leaf tip necrosis (Ltn3) [105]. The frequency of Yr46 gene was found to be 19.1% among wheat genotypes. Genotypes MACS5044, DBW219, K1317, DBW220, HS580, HPBW02, UP2954, WH1215, VL1009, UP2955, VL3002, VL3010, VL3011 and VL3012 were found to be carrying Yr46 and exhibited resistant to moderate field response. Yr46 is widely distributed in older, tall wheat landraces from Punjab regions in India and Pakistan before modern wheat cultivars were grown in this region and was effective in field tests in Pakistan, India, Mexico and Australia and displayed durable resistance [106]. Yr40 and Lr57 is the same gene with pleiotropic effect [84]. Twenty genotypes (29.4%) HI8774, HS626, TL3007, HI1605, K1317, AKAW4842, DBW179, DBW216, DBW217, VL4001, DBW220, WH1184, HS580, VL1009, VL3002, VL3010, VL3011, DDK1050, WH1310 AND HD3171) indicating likely presence of Yr40.
Non-race-specific Yr59 gene, confers a high level of HTAP resistance (IT 2) and is effective against all tested North American Pst races [26]. Eleven genotypes (WH 542, UP 2954, VL 1009, DDK 1050, TL 3009, RKD 283, PBW 760, PBW 757, PBW 723 and HI 8759) identified with Yr59 can be used to strengthen durable breeding programmes. Yr48, is another QTL which confers APR to yellow rust. Twelve genotypes (17.6%) HI 8759, HI 8774, HPW 433, UAS 459, NW 6094, PBW 621, PDW 344, WH 1184, UP 2955, WH 1310 and Lassik were postulated to carry Yr48. An APR gene imparts durable resistance when pyramided together. Hussain et al. [107] showed that genotypes with combinations of slow rusting genes such as Yr18, Yr29 and Yr30 were high yielding with better resistance. This combination is very interesting as it provides protection against three types of rusts (Leaf rust, yellow rust and stem rust). In our study, out of 68 genotypes, ten genotypes (HI 8759 (d), HPPAU 08, HPPAU 10, HPW 424, HS 580, VL1009, VL3002, VL3010, VL3011 and Lassik) showed the combination of three designated slow rusting/durable genes (Yr18 + Yr29 + Yr30). All these ten genotypes also showed immune response at field testing. Asghar et al. [108] studied the stacking effect of combination of yellow and leaf rust resistance genes Yr18/Lr34, Yr17/Lr37, Yr29/Lr46 and Lr47 in 50 spring wheat genotypes using PCR based markers. Yr18/Lr34 gene or loci was observed in 39 genotypes, Yr17/Lr37 in 43 genotypes, Yr29/Lr46 in 33 genotypes and Lr47 was found in 34 wheat genotypes, respectively which exhibited better response towards rust diseases. Our results also showed similar trend with these findings. Madenova et al. [109] identified one genotype with Lr34/Yr18 genes and two genotypes with complex genes Lr37/Sr38/Yr17 using csLv34 and VENTRIUP/LN2 primers. Zheng et al. [36] demonstrated the significant additive effect of Yr9 + Yr18 gene combination in Chinese wheat genotypes. One genotype, HPPAU 05 was identified with the gene combination of Yr9 + Yr18 displaying effective resistance at field testing.
IB267, the translocation carrying resistance gene Sr50 was transferred from Imperial rye into wheat in chromosome 1BL.1RS and 1DL.1RS [55]. It was found to be effective against stem rust race Ug99 and is being used in various Australian wheat breeding program [110]. In our study, two genotypes (PBW 723 & HPPAU 05) were identified with IB267 translocation, which can be useful for incorporating stem rust resistance in wheat breeding programmes. Yr36 is an HTAP gene which is closely linked to Grain protein content gene (Gpc-B1). Two genotypes AKAW 4842 and HI 8774, along with positive control Lassik were found to be carrying Gpc-B1 allele. Genotype HI 8774 also showed linkage with WKS1 gene. These genotypes can be directly utilised in bio-fortification and durable resistance breeding programme.
Among race specific genes Yr5, Yr10, Yr15, Yr24 and Yr26 are still protective against current predominant races in India. Breeding lines containing these genes can be utilised directly in resistance breeding. By analysing the molecular evaluation data, one genotype (VL 3010) was identified with Yr5, Yr10, Yr15 and Yr26 genes. Six genotypes (VL 3011, HS 626, K 1317, WB 2, WH 184 and HS 580) were identified positively for Yr5, Yr10 and Yr24/Yr26 genes. Eight genotypes (PBW 725, AKW 4842, DBW 179, DBW 217, DDK 1050, VL 4001, WH 1215 and HPW 424) showed the presence of Yr15 and Yr24/26 genes. All these lines displayed high immune response at field testing. Moreover, the genotypes with multiple Yr genes identified in this study might be useful as parental lines for diversifying Pst resistance sources in wheat breeding. Indeed, Yr15 gene has been combined with Yr24 and Yr5 in all the commercial varieties in California covering 14% of the acreage of common wheat and proved to be very useful in controlling the yellow rust epidemics in California. The stacking of genes also has not shown any negative traits associated with the introgression of either of these genes [96]. Several combinations of Yr gene such as (Yr5, Yr9, Yr18, Yr26, Yr29), (Yr10, Yr17, Yr18, Yr26, Yr29), (Yr10, Yr17, Yr18, Yr26, Yr29) and (Yr5, Yr10, Yr18, Yr26, Yr29) was also reported by Begum et al. [111] in Pakistani wheat lines.
Out of 53 parental lines showing field resistance, 9 lines PBW723, HS622, HS623, PBW757, PBW760, TL3006, UP2955, Lassik and WH542 showed a resistant response to yellow rust races but molecular studies indicates these genotypes do not carry Yr5, Yr10 and Yr15. Genotype, PBW 757 showed least number of Yr genes but exhibited immune response in field testing. For genotypes, exhibiting positive reaction in the field for rust resistance but not showing likely presence of major rust resistance Yr5, Yr10 and Yr15 genes, must be having other effective combination of ASR and APR genes or other effective genes which could not be detected with the linked markers used in the study or lack the corresponding allele which could be as a result of genetic recombination [112].
5 Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Compliance with ethical standards
Disclosure of interest
The authors declare that they have no competing interest.
Acknowledgments
We sincerely acknowledge Chaudhary Charan Singh Agricultural University, Hisar, Haryana, India for providing financial support and the facilities to carry out this research work.


