1 Introduction
Environmental pollution is largely blamed for having a negative influence on human health, particularly male infertility, and for being responsible for the increasing incidence of marital infertility in some parts of the world [1,2]. It is therefore important to identify the environmental factors that could alter the male reproductive function to understand why male human reproductive disorders like cryptorchidism, hypospadias, low sperm counts and testicular cancer may be increasing in incidence in many countries in the last few decades [3].
Male infertility can be induced by multiple mechanisms and, in addition, many different factors play a role, for example, in the alteration of sperm parameters [3,4]. Sperm morphology has long been considered an important factor for evaluating semen and sperm quality. Unfortunately, manual assessments remain problematic because there can be large variations among technicians and laboratories owing to subjective evaluation of sperm morphology. Thanks to computer-aided sperm morphometry analysis (CASMA), automated analysis of rat sperm morphology and morphometry may be useful tools for quantification of the effects of reprotoxicants on sperm morphology [5].
Epidemiological, clinical, and experimental arguments suggest that idiopathic male infertility results from harmful effects caused by pollutants and that they may act by disrupting the epigenetic mechanisms [6,7]. Indeed, the role of epigenetic modifications, like DNA methylation, is involved in the regulation of spermatogenesis in testis and in sperm function [8,9]. Other environmental factors such as ionizing radiation could alter the DNA methylation in sperm, modifying the gene expression and inducing male infertility [10]. In addition, an important question may be raised in regard to whether such modifications in methylome and gene expression are transmitted to the next generation. An increasing numbers of environmental pollutants (like endocrine disruptors compounds) are involved in epigenetic transgenerational reproductive disorders, confirming the need for performing multi- and transgenerational experimental studies [11].
In previous experimental studies, chronic low-dose exposures to uranium, a water environmental pollutant [12], were associated with male reproductive defects [13,14] and modifications of DNA methylation in gonads [15]. Elmhiri et al. demonstrated that chronic low-dose exposure of uranium affects the global methylation profile in the gonads and that this profile has been conserved across generations. The aim of the current investigation was to establish, for the first time, whether chronic exposure to Natural Uranium (NU) could (i) modify the morphometric sperm parameters, (ii) induce epigenetic effects (DNA methylation) in spermatozoa of rats after multiexposure of the F0, F1, and F2 generations.
2 Material & methods
2.1 Experimental design and uranium contamination
All experimental procedures were approved by the Animal Care Committee of the Institute of Radioprotection and Nuclear Safety (IRSN, France) and complied with French regulations for animal experimentation (Ministry of Agriculture Act No. 87-848, 19 October 1987, modified 20 May 2001).
NU (Mc Arthur) was obtained from CERCA (Pierrelatte, France). Uranyl nitrate hexahydrate (UO2(NO3)2·6H2O). The drinking solution was prepared to obtain a final uranium concentration of 40 mg·L−1 in mineral water (daily uranium intake dose: 1 mg/rat/day) [16]. The specific activity of the NU was 2.42 × 10+4 Bq·g−1, and its isotopic composition was 238U ∼ 99.307%, 235U ∼ 0.688%, and 234U ∼ 0.005%. This concentration of NU is not nephrotoxic [12] and three times higher than the highest uranium concentration of 12.4 mg·L−1 naturally, found in well water in Finland [17], to half of the WHO 2011 drinking-water guideline for uranium, defined as equal to 0.030 mg·L−1 [18]. All control groups of rats received ad libitum uncontaminated mineral drinking water Evian® (Évian-les-Bains, France).
Each experimental group was composed of a number of 20 rats. This number has been calculated to highlight low biological effects associated with low-dose exposure with a sufficient statistical power.
Outbred Sprague–Dawley, 12-week old and 16-day pregnant, female rats were obtained from Charles River Laboratories (L’Arbresle, France). They were housed individually and maintained in a 12 h light/12 h dark cycle (regular cycle) at 21 °C and 50% humidity, with access ad libitum to a standard rodent pellet diet and water until birth (Fig. 1).
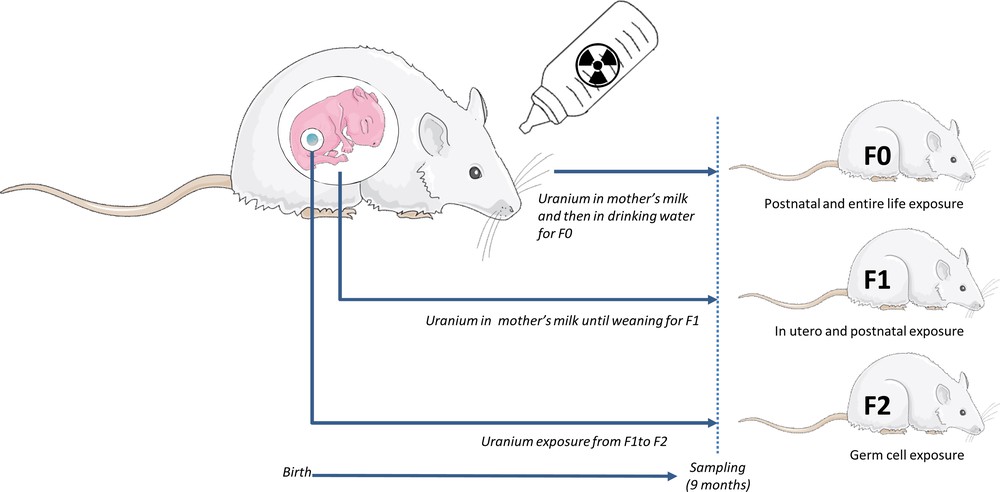
A multi-generational protocol of exposure, i.e. three generations of rats (F0, F1, and F2) males and females (n = 20). F0 rats were exposed with NU from birth through their mother's milk and then through from drinking water for 9 months (control animals drank uncontaminated mineral water). F1 rats were contaminated in utero and through their mother's milk (F0) until weaning. After weaning, they drank uncontaminated mineral water. The last F2 generation received only mineral water. The last generation was exposed to uranium only from parental (F1) germ cells.
The first generation (F0) of the treated group was chronically exposed to NU through drinking water from their birth to the age of 9 months. During weaning (three first weeks of life), rats (F0) were mostly exposed to NU through lactation (human offspring receives approximately 5% of the mother's daily uranium dose [19]) and through the contaminated drinking water. At weaning, male and female (F0) of different bearing were sorted and randomly housed together. Each experimental group (control vs contaminated) was itself divided to obtain breeding and non-breeding subgroups. Finally, four F0 experimental groups were thus created. F0 male and female rats were contaminated via drinking water supplemented with NU. Rats of the breeding group were mated after 6 months of exposure (a sufficiently long time to chronically expose rats to NU). To compose the next generation F1, one male was housed and mated with two females during 5 days as recommended [20]. Rats of the non-breeding group were exposed to NU until the age of 9 months (Fig. 1).
The next generation (F1), was exposed to NU in utero (sensitive fetal-developmental windows [21]) and after birth through the mother F0 milk. At weaning, the protocol of contamination was stopped and all rats received uncontaminated mineral drinking water ad libitum. Males and females F1 of different bearing were sorted and randomly housed (control vs contaminated and themselves divided to obtain breeding and non-breeding subgroups.). Breeding groups F1 were mated following the same conditions as previously done for F0 and non-breeding groups housed until the age of 9 months. Indeed, the last generation F2 was contaminated only from the parental germ cells F1. All animals were euthanized at 9 months of age (Fig. 1).
At 9 months, rats were anaesthetized by inhalation of 5% isoflurane (Abbot France, Rungis, France) and euthanized by an intracardiac puncture. The paired epididymis were removed and weighed after releasing adipose tissues. The relative weight of organs was estimated by calculating the ratio of organ weight to body weight The organs were deep-frozen in liquid nitrogen, and stored at −80 °C.
2.2 Sperm isolation and staining
Directly after sacrifice, the epididymides were dissected free from blood vessels, fatty tissue and other connective tissue, and they were placed in a Petri dish containing 5 mL of a solution of M199 medium (Sigma-Aldrich, St Louise, MO, USA) at 37 °C. The proximal caudal portion of each epididymis was isolated and placed in 5 ml of fresh medium in 6 well plates at 37 °C. A short length of “cleaned” epididymal tubule was removed by using a scalpel.
Sperm flowed freely from the ends of the transected tubule into the medium and a cloud of sperm quickly formed. Media containing sperm were collected from the edges of the sperm ‘cloud’ using a positive displacement pipette (15 μL). For staining sperm, the sperm suspension was placed on a slide and a sperm smear was made by dragging the fluid behind another slide while spreading it at an angle of 45° across the slide. After air drying at room temperature, slides were fixed by immersing them horizontally into SpermBlue® fixative (Microptic SL, Barcelona, Spain) for 10 min, and stained thereafter with the SpermBlue® stain for 20 min, following a similar immersion technique [22]. Stained slides were then gently submerged in distilled water for 3 s to remove excess stain, and allowed to dry in the air at an angle of 60–80°. The slides were finally mounted using Eukitt (O. Kindler GmbH, VWR, France).
2.3 Sperm morphology and morphometric analysis
Sperm morphology and morphometry analyses were performed using the CASMA Sperm Class Analyser system (SCA®; Microptic SL) version 5.4 software and using methods previously described [5]. Analysis used bright field optics employing a 60 × objective, i.e. 600 × magnification, and a blue filter on a Nikon E200 microscope. One hundred sperm/animal were analyzed (N = 10/per group). All spermatozoa were measured randomly, to exclude any bias in favor of a specific morphological form. Images were captured digitally using a Basler 780–75 gigaethernet camera (Basler Microscopy Camera, Germany). The SCA system's morphology module RatTox (Microptic S.L., Barcelona, Spain) automatically analyzes morphometric dimensions of the sperm head and middle-piece (MP). For head morphometry, head length (ARC), width, surface area, perimeter, angle, and roughness (which varies with a range between 0 and 1, and expresses the degree of resemblance of the sperm head to a circle) were measured. Measurements of the MP included width and angle of insertion of the flagellum to the head. The chord length, i.e. the distance from the anterior tip of the acrosome to the posterior part of the head, was measured. Linearity is a derivative expressed as a percentage and refers to ARC/chord × 100 (Table 1).
Morphometric variables of the sperm head measured automatically by SCA®.
| Parameters | Formula |
| Arc (μm) | L |
| Area (μm2) | A |
| Perimeter (μm) | P |
| Chord (μm) | C |
| Linearity (%) | L/C |
| Roughness | 4π(A/P2) |
To determine the morphologically normal from the morphologically abnormal sperm in animal species, we used the new approach to determine cut-off points for normal vs abnormal rat sperm previously described [5]. Our method was based on grouping the different morphometric measurements for each percentile, e.g., for each morphometric parameter. The minimum and maximum values for each specific morphometric parameter were derived from the percentile intervals (5 − 95%) (Table 2). The total morphologically abnormal sperm indicating the sum of abnormal head and MP was also determined. The percentage of abnormal sperms was also calculated if sperm has at least one, only one or more than two abnormalities in the head, in MP or in both compartment (head and MP).
Minimal and maximal values for normal head and MP properties selected to determine normal vs. abnormal sperm under the configuration set-up of the SCA 5.4 system using a 5–95% confidence interval.
| Sperm head and middle-piece parameters | Minimum value | Maximum value |
| H Length = Arc (μm) | 18.48 | 23.62 |
| H Width (μm) | 0.84 | 1.51 |
| H Area (μm2) | 14.12 | 21.64 |
| H Perimeter (μm) | 38.95 | 49.13 |
| H Angle (degrees) | 40.63 | 84.16 |
| H Linearity (%) | 49.06 | 66.67 |
| H Chord (μm) | 10.44 | 13.53 |
| H Roughness | 0.09 | 0.14 |
| MP Width (μm) | 0.28 | 0.79 |
| MP Angle (degrees) | 1.44 | 29.54 |
To determine the sperm concentration, the epididymis was entirely minced and homogenized. For each animal, the sperm concentration was determined in triplicate using Malassez. Counting results are expressed as the total number of sperm cells per mL of epididymis. Sperms were stored at–80 °C for subsequent experiments.
2.4 Epigenetics analysis
2.4.1 DNA extraction
Isolation of genomic DNA from sperm was optimized according to QIAamp DNA Mini Kit (QIAGEN, France): 100 μL of semen (5.106 cells) was transferred into a new 1.5 mL tube and then 100 μL of buffer X2 (20 mM Tris–Cl pH 8.0; 200 mM NaCl; 20 mM EDTA; 4% SDS), 80 mM DTT, 250 μg/mL proteinase K were added and gently mixed. The samples were incubated at 55 °C for 1 h. Thereafter, 200 μL of buffer AL and 200 μL of ethanol (100%) were added and mixed. The elution of DNA in 50 μL was finally obtained following the tissue protocol (steps 5 to 8) in the QIAamp DNA Mini Kit (QIAGEN, France). The samples were stored at −20 °C until further analysis.
2.4.2 Global DNA methylation
Global DNA methylation of epididymal sperm was detected using an ELISA reaction with a monoclonal antibody sensitive and specific for 5-methylcytosine (5-mC) and a horseradish peroxidase conjugate as secondary antibody (5-mC DNA ELISA Kit, Zymo Research, Irvine, CA, USA). The level of 5-mC in DNA is reported as the amount of methylated cytosine relative to the cytosine genomic content (%). An aliquot of the sample containing 100 ng DNA was added to the 5-mC coating buffer and brought to a final volume of 100 μL. All samples and controls were denatured at 98 °C for 5 min in a thermocycler and immediately cooled on ice for 10 min. Controls and samples were added to the ELISA plate and incubated at 37 °C for 1 h. After discarding the coating buffer, the wells were washed three times with the 5-mC ELISA buffer and incubated again with 200 μL of 5-mC ELISA buffer at 37 °C for 30 min. The buffer was discarded from the wells and the samples were incubated with the antibody mix at 37 °C for 1 h. The antibody mix consisted of the 5-mC ELISA Buffer, anti-5-methylcytosine and the secondary antibody in a ratio 1:2000:1000. After incubation, the antibody mix was discarded and a 100 μL HRP were added to each well. The absorbance was measured in duplicate at 405 nm using TECAN reader (Infinite® 200 PRO NanoQuant Plate Reader). The results are expressed as % 5-mC per total C.
2.4.3 RNA extraction and qPCR
Total RNA was extracted from sperm, using mirVana™ miRNA Isolation Kit (Ambion, cat.no.1560). The NanoDrop apparatus (ThermoFisher Scientific, Cergy-Pontoise, France) was used for determining the concentration of RNA ng/μL. One microgram of total RNA was reversely transcribed using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Courtabœuf, France) according to the manufacturer's instructions. Real-time qPCR was performed with QuantStudio 12 K Flex Real-Time PCR System (ThermoFisher Scientific, Cergy-Pontoise, France) using TAQMAN (Applied biosystems), to analyze the mRNA levels of demethyls genes TET2 (Rn01522037_m1), TET3 (Rn01425643_m1), TDG (Rn00821513_m1), with three internal controls (GAPDH (Rn01775763_g1), Ywaz (Rn00755072_m1) and β2 M (Rn00560865_m1)) and using SYBR Green® (Applied Biosystems) to analyze the mRNA levels of methyltransferases: DNMT1, DNMT3a, DNMT3b, DNMT3l with three internal controls HPRT, β2M, and ACTB (Table 3); relative changes in genes mRNA expression in sperm were calculated using e-ΔCT method and three internal controls [23,24]. All RTqPCR results are expressed as mean standard deviation, and compared with expression levels of the control group.
Sequence of primers set for gene expression studies in sperm.
| Genes | Sequences |
| DNMT1 | 5’-CGGCGGAGGTGTCCTAACTTGGC-3’ 5’-GGGTGACGGCAACTCTGGTA-3’ |
| DNMT3a | 5’-CGGTAGCGCCTCTTCTTTGAGTTCTAC-3’ 5’-GCGATCATCTCCCTCCTTGG-3’ |
| DNMT3b | 5’-GGGCCGCTACCACGTTCAGG-3’ 5’-AGGGCCGTCCTGGCTCAAGT-3’ |
| DNMT3l | 5’-GCTTTGACGGTGGCGAGAA-3’ 5’-TCTGCAAGAACTCGACCACAATC −3’ |
| HPRT | 5’-GCTCGAGATGTCATGAAGGAGA-3’ 5’-TCAGCGCTTTAATGTAATCCAGC-3’ |
| β2M | 5’-ACATCCTGGCTCACACTGAA-3’ 5’-ATGTCTCGGTCCCAGGTG-3’ |
| ACTB | 5’-TTCAACACCCCAGCCATGT-3’ 5’-TGGTACGACCAGAGGCATACAG-3’ |
2.4.4 Statistical analysis
Results are expressed as mean ± standard deviation (SD), Student's t-test was routinely performed for statistical analysis of data, and was replaced by Mann–Whitney Rank Sum Test when the equal variance test failed (determined by Sigmaplot Stat software). The Fisher exact test was used to compare fertility index and a quasi-Poisson regression was conducted for the anomaly rates modeling.
Since the epididymis data consist of a paired observation within the same rat, a reasonable assumption is that these observations are correlated and therefore, statistical methods that recognize and account for the correlation of observations within a subject are appropriate. Thus, a generalized estimating equations (GEE) regression [25] was conducted using the Zelig Package of the R software [26].
Differences were considered statistically significant when p ≤ 0.05.
3 Results
3.1 Fertility index & epididymis relative weights
The multigenerational exposure to uranium had no effect on the number of offspring born, male/female sex ratio, or pup mortality in any of the F0, F1, and F2 generations. The average litter size was constant between the control and U-exposed generations (data not shown). However, the pregnancy rate was significantly decreased (55%) in the F1 U-exposed generation (35%) in comparison to the pregnancy rate of the F0 U-exposed generation (70%) (Table 4). The neonatal mortality rate was less than 5% for both control and U generations (data not shown). The epididymis/body weight ratio between the control and U generations was statistically different only in the F1 generation, in which it decreased to −15.4 ± 0.6% in comparison to the control group (Table 4).
Pregnancy rate and epididymis relative weight were compared in the F0, F1, and F2 generations.
| Parameters | F0 | F1 | F2 | |||
| Control | U | Control | U | Control | U | |
| Pregnancy rate | 70% | 70% | 67% | 30%a | No reproduction | |
| Epididymes Relative weight g/100 g body weight |
0.115 ± 0.012 | 0.113 ± 0.023 | 0.118 ± 0.011 | 0.108 ± 0.027>remove bolda | 0.115 ± 0.020 | 0.122 ± 0.029 |
a p < 0.05. N = 17–20 per group.
3.2 Morphometric parameters and sperm morphology
Chronic exposure to uranium did not alter the sperm concentration in the F0, F1, and F2 generations as compared to the control group (data not shown). However, morphometric analysis of sperms showed that uranium affected sperm morphology from the F0 generation. The angle of the MP was reduced from 14.85 ± 2.84° to 12.02 ± 3.33° in the F0 U-generation. The area of the head was statically increased in the F1 U-generation (Table 5). Using the cut-off values, we determined the percentage of abnormal sperm for each morphometric parameters (Table 2, Figs. 2 and 3). Furthermore, it was found that uranium significantly induced sperm abnormalities for four parameters in F0 U-exposed generations as ARC, area and perimeter of the sperm head, and angle (for the MP) (Fig. 2A). The percentage of abnormal sperms head (angle, linearity), and the MP width from the F0 U-generations were significantly different between control and U-exposed groups (Fig. 2B). In F2-U generation, there is only an increase in abnormal sperm for the area of the sperm head and the angle of the MP (Fig. 2C). The percentage of sperm with at least one abnormality in the head, and at least one or only one abnormality in the MP was also increased in the F1 and F2 generations, but no difference was observed in F0 (Fig. 3D to F). The percentage of sperm with one abnormality in the head and in the MP was modified in the last generations (F1 and F2) (Fig. 3D and E). In addition, we have detected that the percentage of total abnormal sperm was significantly higher on the F1 U-exposed generations, and that there was a significant higher percentage of sperms with one abnormality both in the head and in the MP (Fig. 3E).
Mean value for head and MP properties selected to determine abnormal parameters in each generation.
| Sperm head and middle-piece parameters | F0 | F1 | F2 | |||||||
| Control | U | p-value | Control | U | p-value | Control | U | p-value | ||
| Head | Length = Arc (μm) | 21.232 ± 0.273 | 21.457 ± 0.538 | 0.310 | 20.972 ± 0.641 | 21.381 ± 0.416 | 0.122 | 21.392 ± 0.349 | 21.315 ± 0.572 | 0.736 |
| Width (μm) | 1.062 ± 0.093 | 1.021 ± 0.075 | 0.345 | 1.143 ± 0.045 | 1.174 ± 0.035 | 0.115 | 1.223 ± 0.131 | 1.165 ± 0.139 | 0.381 | |
| Area (μm2) | 16.834 ± 1.248 | 16.961 ± 0.858 | 0.817 | 16.640 ± 0.979 | 17.946 ± 1.209 | 0.019a | 18.214 ± 1.516 | 17.286 ± 1.253 | 0.176 | |
| Perimeter (μm) | 44.353 ± 0.597 | 44.796 ± 1.114 | 0.339 | 43.841 ± 1.243 | 44.707 ± 0.823 | 0.095 | 44.912 ± 0.896 | 44.595 ± 1.086 | 0.509 | |
| Angle (degrees) | 64.710 ± 2.025 | 64.731 ± 2.912 | 0.987 | 64.006 ± 1.987 | 64.007 ± 2.707 | 0.948 | 64.663 ± 2.490 | 65.763 ± 2.124 | 0.328 | |
| Linearity (%) | 57.449 ± 1.337 | 57.447 ± 2.151 | 0.998 | 57.263 ± 0.15 | 56.722 ± 1.586 | 0.355 | 56.486 ± 1.756 | 56.764 ± 1.482 | 0.722 | |
| Chord (μm) | 12.210 ± 0.211 | 12.339 ± 0.247 | 0.281 | 12.051 ± 0.391 | 12.128 ± 0.265 | 0.626 | 12.096 ± 0.353 | 12.101 ± 0.333 | 0.974 | |
| Roughness | 0.108 ± 0.006 | 0.107 ± 0.003 | 0.587 | 0.110 ± 0.002 | 0.114 ± 0.005 | 0.111 | 0.114 ± 0.006 | 0.110 ± 0.008 | 0.247 | |
| Middle-piece | Width (μm) | 0.568 ± 0.061 | 0.590 ± 0.078 | 0.555 | 0.540 ± 0.050 | 0.579 ± 0.088 | 0.153 | 0.554 ± 0.046 | 0.533 ± 0.036 | 0.305 |
| Angle (degrees) | bold> | 12.025 ± 2.337 | 0.047a | 12.880 ± 1.926 | 13.852 ± 2.486 | 0.352 | 12.695 ± 2.153 | 11.088 ± 2.043 | 0.127 |
a p < 0.05. N = 8–10 per group.
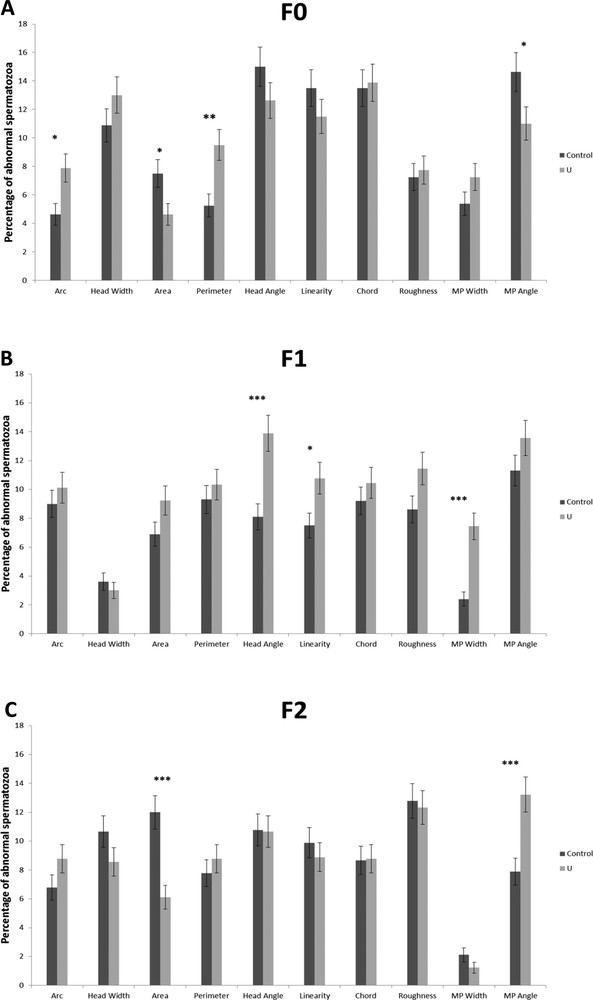
The percentage of abnormal sperm for each morphometric parameters in the F0, F1, and F2 generations (A, B, and C, respectively) (mean ± SD). There was a significant difference between the control group and the NU40 group (*p < 0.05; **p < 0.005; *** < 0.001). N = 8–10 per group.
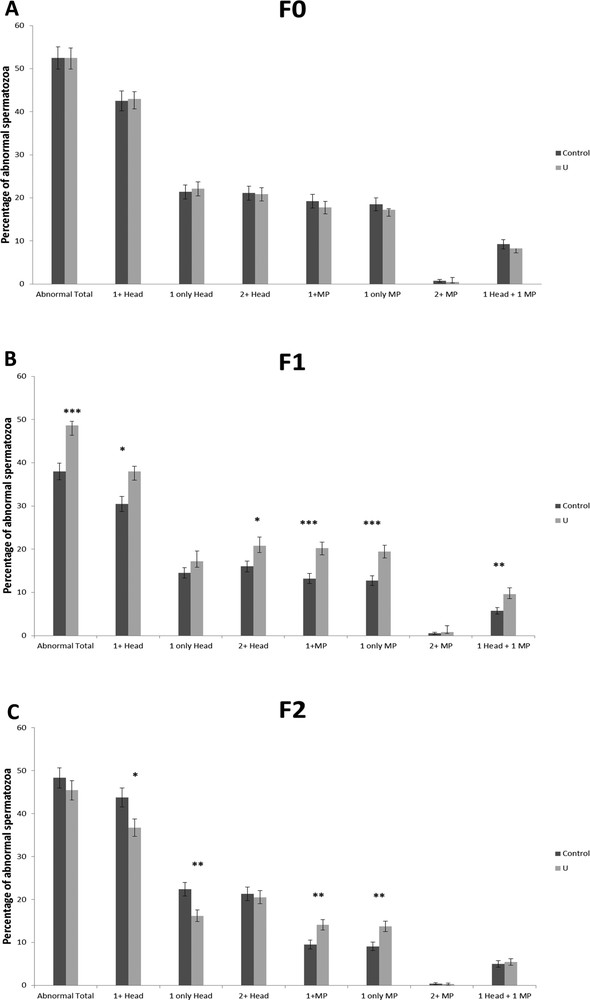
The percentage of abnormal sperm for each compartment (head and MP) in the F0, F1, and F2 generations (A, B, and C, respectively) (mean ± SD). Total morphologically abnormal sperm indicating the sum of abnormal head and MP was determined. The percentage of abnormal sperms was also calculated if sperm has at least one, only one or more than two abnormalities in the head, in MP or in both compartments (head and MP) for the F0, F1, and F2 generations. There was a significant difference between the control group and the NU40 group (*p < 0.05; **p < 0.005; *** p < 0.001). N = 8–10 per group.
3.3 Global DNA methylation & gene expression of DNA methyltransferases and demethylation enzymes
The level of global semen DNA methylation was not significantly modified in the F0 and F1 generations (Figs. 4A and 4B). Nevertheless, DNA hypomethylation was observed (−25%) in sperm from the F2 generation (p < 0.005) (Fig. 4C).
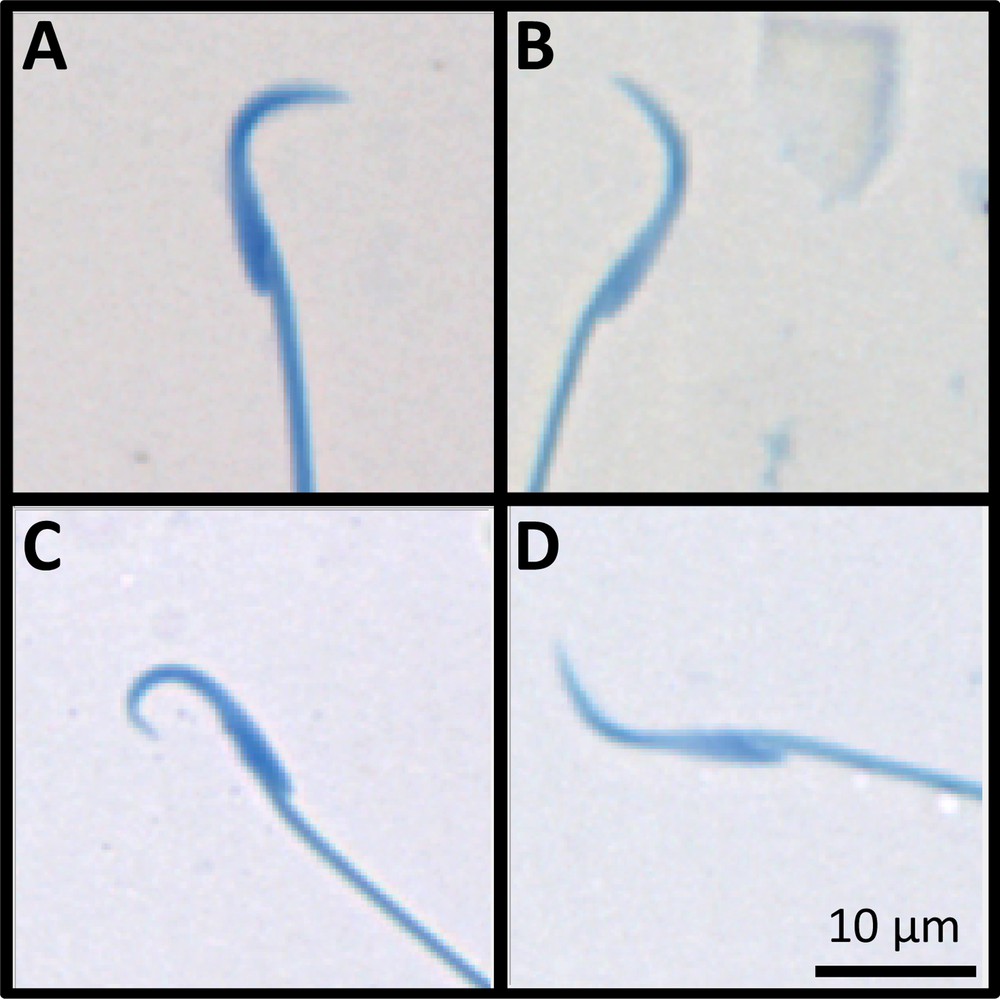
Examples of abnormal stained rat sperm, automatically analyzed for morphology and morphometric parameters. A: Control sperm (F1 generation); B: F0 U-exposed sperm with abnormal head parameters in the angle, chord and linearity (low values); C: F1 U-exposed sperm with abnormal head parameters in the angle, chord and linearity (upper values); D: F2 U-exposed sperm with abnormal head parameters in the angle, chord and linearity (low values).
mRNA analysis showed that there were no significant difference in transcripts levels of methylation enzymes, DNMT1, DNMT3a, and DNMT3b, between control and U-exposed groups in the F0, F1 and F2 generations (Fig. 4D to F). However, DNMT3L was down-regulated (p < 0.05) in comparison with controls in the F0 generation (Fig. 4D).
The analysis of the demethylated genes (TET2, TET3 and TDG) in the F0 and F1 generations showed that there was no significant difference between control and U-exposed groups (Fig. 4D to F). But, the expression rate of TET2 and TET3 in the F2 generation decreased significantly (p < 0.05) (Fig. 4F).
4 Discussion
Currently, it is well accepted that a change in some epigenetic mechanisms marks such as DNA methylation after exposure to certain pollutants could be transmitted through generations with the appearance of deleterious effects such as infertility, reproductive dysfunctions [6,7,11]. In the field of chronic exposure to radionuclides, very little scientific data are available. Our experimental model allows us to study the effects of uranium exposure on morphometric parameters and epigenetics in rat sperms after multigenerational exposure (Fig. 5).
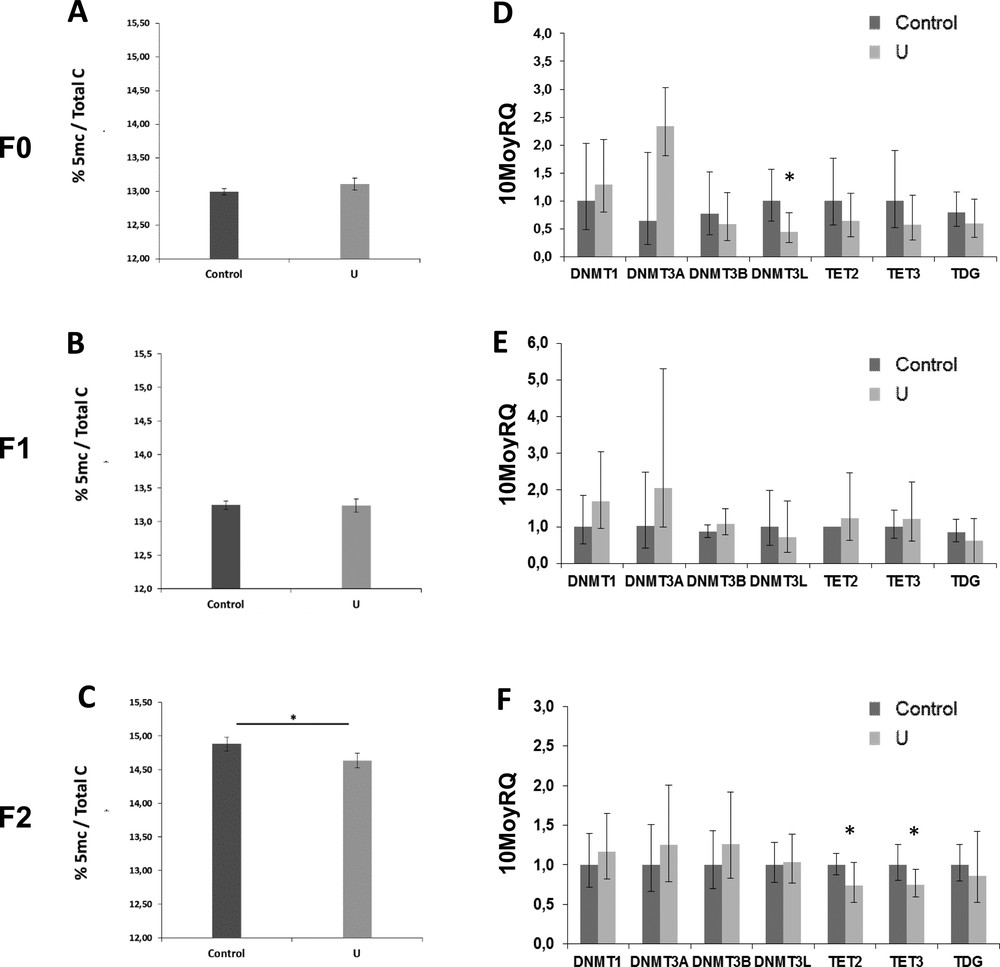
Percentage of global DNA methylation assessed with ELISA (mean ± SD) in the sperm of the F0, F1, and F2 generations (A, B, and C respectively). mRNA expressions of DNA methyltransferase specific genes (DNMT1, DNMT3a, and DNMT3b) and DNA demethyltransferase specific genes (TET2, TET3, and TDG) in the sperm of the F0, F1, and F2 generations (D, E, and F respectively). All results are expressed as mean ± SD. There was a significant difference between the control group and the NU40 group (*p < 0.05; **p < 0.005). N = 8–10 per group.
First, we detected that the pregnancy rate was affected in the F1 U-generation. This result could be explained by the exposure period of animals, i.e. from the fetal life to their weaning, and which includes the time of male sex determination and the development of the male reproductive organs. To study the effects of U on rat sperm, we explored the rat sperm morphometry and morphology in rats in the F0, F1, and F2 generations. Using automated sperm morphology analysis, we compared ten sperm morphometric parameters between control and U-exposed groups in each generation. Interestingly, the analysis shows firstly that the determination of the cut-off values was reproducible with those obtained in Sprague-Dawley rats in the previous study using the CASMA [5]. We detected that U exposure could affect two compartments of the sperm morphology, i.e. the head and the middle-piece in the F0, F1, and F2 generations, but without common parameters between each generation. The percentage of total abnormal sperms was significantly higher only in the F1 U-exposed sperms, when the animals were exposed during their fetal life from weaning, contrasting with F0 (from birth until 9 months of age) and F2 (as germ cell lines of F1) generations. These results confirm the higher susceptibility of rats to U during the fetal and postnatal exposure and the effects of multigenerational exposure to U, which was previously suggested [14,27,28]. Regarding the Developmental Origins of Health and Disease (DOHaD) hypothesis [29], the effects of the exposure to U may be involved in reproductive disorders. Further experiments should be performed in our multigenerational exposure to identify the adverse effects on sperm motility and testicular functions, where spermatogenesis takes place.
No epidemiological studies showed sperm defects in a U-exposed population [30,31]. Only modifications in the sex ratio of offspring or in the testosterone level were reported [32,33]. Experimental studies with chronic exposure showed abnormalities of the sperm head in adult male rats exposed to 160 mg·L−1 and in two generations of mice exposed to 64 mg·L−1 [34,35]. Until recent years, little attention had been paid to the potential adverse effects of uranium (including natural and depleted uranium) exposure on human reproduction.
In recent years, numerous studies have described the impact of environmental exposures on the reproductive system associated with epigenetic modifications [6,7,11]. In our study, we have also observed epigenetic modifications induced by chronic exposure to uranium. Indeed, contrary to a recent study that showed that DNA was hypermethylated in testis [15], in a very surprising and interesting way we observed an overall hypomethylation in the sperm of rats exposed to NU and particularly observed from the last generation (F2). We analyzed for the first time the mRNA expression of DNA methyltransferase and demethylase enzymes (DNMT, TET and TDG) in rat sperm, already described in mice and humans [36–38]. We observed only a decrease in DNMT3L expression for the F0 generation. DNMT3L has not catalytic activity but it interacts with DNMT3A and DNMT3B to stimulate their activity [38]. Interestingly, DNMT3L knockout phenotype is related to abnormal maternal imprinting and male infertility [39]. In the F2 generation, only the gene expression of the enzymes of the TET family (TET2 and TET3) was modified. In ejaculated spermatozoa from humans with oligozoospermia and/or asthenozoospermia, the expression of these TETs is reduced, suggesting their pivotal role in male fertility [37]. Moreover, together, the mRNA profiles are insufficient to explain the hypomethylation of the global sperm DNA observed from the F2 generation. The identification of differential methylated regions needs to be realized using targeted methylation tools as already used [7,11].
Our study focused on male reproductive disorders, but females could also be involved in the low pregnancy rate observed in F1. A previous study has already demonstrated the modification of the DNA global methylation rate in the ovary of the female exposed in the same multigenerational model [15]. Moreover, in order to explore the pregnancy rate in the F1 U-exposed generation, females that were not pregnant were analyzed regarding their estrus cycle. We detected that the estrus phase was reduced and the diestrus phase was significantly longer in the U-exposed females (data not shown). These results suggest the involvement of both sexes in the multigenerational effects of U exposure. Female reproductive function should be more explored in subsequent studies.
5 Conclusion
Thanks to our multigenerational exposure, we showed that uranium-induced abnormalities in sperm morphometry and morphology occur, both in the head and the middle piece. These abnormalities were observed in each generation and were more deleterious in the F1 U-exposed generation, suggesting that exposure to U during fetal life may therefore have more adverse effects on reproductive parameters. Consequently, the decrease in the pregnancy rate to generate the F2 offspring could be the consequences of these morphological defects in the sperm. Epigenetic change may be one possible consequence of the effects of U on sperm. Indeed, our results show epigenetic effects as DNA global methylation (overall sperm hypomethylation) observed in F2. These results raise the question of the biological significance of the epigenetic germinal fingerprints to subsequent generations. There is an increasing proof about a strong link between errors in sperm DNA methylation and male hypofertility [6,40]; this study provides also new insights into the sperm morphometry and the epigenetic change induced after uranium exposure and their persistence over generations.
Disclosure of interest
The authors declare that they have no competing interest.
Funding information
This study was part of the ENVIRHOM research program supported by the Institute for Radioprotection and Nuclear Safety (IRSN).
Acknowledgement
The authors thank M. Voyer, A. Sache, M. Loiseau, and M. Guischet who took good care of the animals. The authors would like to acknowledge Souad Nebchi for proofreading of the manuscript.


